CD Genomics provides accurate and reliable HLA typing service based on next-generation sequencing (NGS) technology that generates unambiguous, phase-resolved HLA sequencing results in a single assay.
What is HLA
The major histocompatibility complex (MHC), also known as the major histocompatibility complex gene, is a gene family present in the genomes of most vertebrates and is closely associated with the immune system. MHC has two types, with the first type (MHC class I) processing proteins that have been broken down inside the cell (such as viruses). MHC class I is widely distributed and is present on the surface of all nucleated cells in the body. On the other hand, the second type (MHC class II) is primarily expressed on the surface of certain immune cells. Its function is to present fragments formed when invaders are engulfed externally and processed by lysosomes. MHC binds to these fragments and presents them on the cell surface for recognition by T cells (antigen presentation).
In humans, the Major Histocompatibility Complex (MHC) is known as the Human Leukocyte Antigen (HLA), with its genes located on the short arm of chromosome 6 within a range of 3600kb, encompassing 224 genetic loci. Each locus contains numerous alleles, making it the most complex known genetic polymorphic system in the human body, with tens of thousands of gene polymorphisms. The IPD-IMGT/HLA Database is a comprehensive repository of HLA alleles, currently housing over 30,000 alleles. Around 17 × 107 genotypes have been identified, with almost no individuals in the population, other than identical twins, sharing the same HLA profiles, earning HLA the moniker of a "genetic ID card."
Aside from organ transplantation, HLA antigens are also implicated in various diseases such as autoimmune diseases, infectious diseases, cancer, and adverse drug reactions. As a result, HLA typing finds wide applications in fields such as transplant medicine, disease diagnostics, and cellular immunotherapy.
The Introduction to HLA Typing
HLA genotyping is the identification of the HLA class I and class II gene polymorphisms for individuals, which is indispensable for transplant matching and disease association studies. Unambiguous HLA genotyping is technically challenging owing to high polymorphism in various genomic regions. The development of NGS has changed this landscape of genotyping. High-resolution HLA genotyping by using PCR and NGS is uniquely able to address limitations of traditional HLA genotyping and Sanger sequencing assays in patients. It enables robust, simple, high-quality, and high-throughput analysis of the key HLA genes, data can be phased to a minimum of 6 digits. Another advantage is that phasing problem is determined since DNA templates are derived from single molecules.
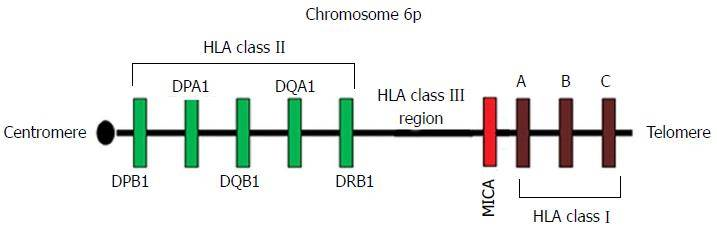 Figure 1. The HLA region in Chromosome 6.
Figure 1. The HLA region in Chromosome 6.
High-quality Next-Generation Sequencing (NGS) methodologies heavily rely on the precise enrichment of the pertinent HLA loci, achieved through either PCR- or bait-based capturing techniques. Initially, the target DNA is enriched, followed by the sequencing of randomly fragmented DNA, culminating in a detailed data analysis. Target enrichment methods encompass bait-based capturing, long-range PCR, or even shorter PCR amplicons spanning the region of interest.
Nevertheless, NGS in isolation may not offer a comprehensive understanding of the complete HLA gene haplotypes. To address this limitation, we present an all-encompassing workflow for sequencing HLA class I and II genes, leveraging long-read sequencing technologies, notably SMRT and Nanopore sequencing. This cutting-edge approach facilitates precise HLA typing and the discernment of allele-specific HLA expression profiles.
Advantages of Our HLA Typing Service
- High resolution and 100% accuracy
- Flexible testing sites: such as HLA class I (A, B, C) and HLA class II (DQB1, DQA1, DPA1, DPB1, DRB1, DRB3, DRB4, DRB5)
- Capture sequencing enabling a cost-effective, fast, and high-throughput manner
- Strict quality control and rich practical experience
- Sample-to-Report Solution
- Our HLA typing service has been widely applied in organ transplantation, population evolution, gene therapy, as well as immunological disease and cancer studies.
Applications of HLA Typing
- Suitable for the study of transplant immune-related diseases or specific conditions.
- Appropriate for high-resolution typing in clinical transplantation.
- Relevant for HLA research projects and investigating genetic variations.
HLA Typing Workflow
Our highly experienced expert team executes quality management following every procedure to ensure comprehensive and accurate results. Our HLA typing workflow is outlined below, including DNA isolation, HLA gene capture, library preparation, high-throughput sequencing, and bioinformatics analysis.
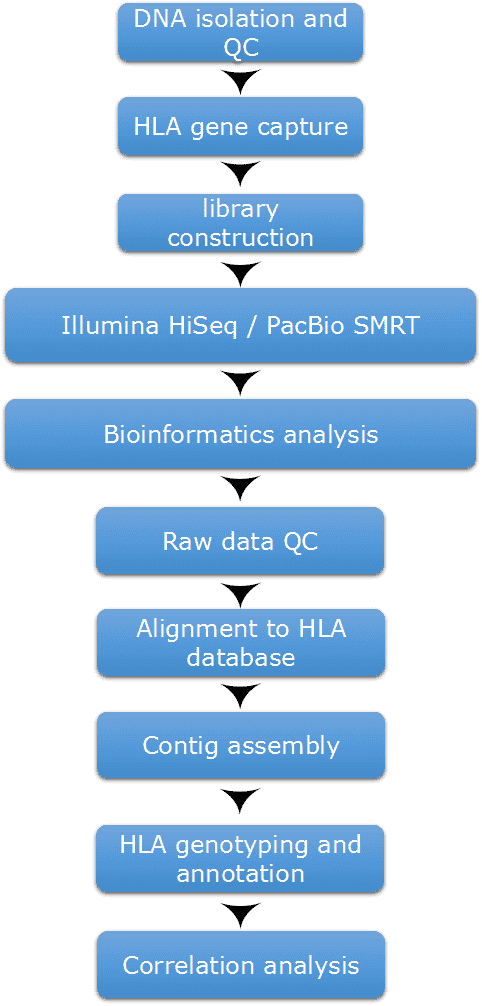 Figure 2. Our workflow of HLA typing.
Figure 2. Our workflow of HLA typing.
Service Specifications
Sample Requirements
|
|
Click |
Sequencing Strategy
|
| Bioinformatics Analysis We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
HLA Typing: Key Types, Testing Methods, and Transplant Significance
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in HLA Typing for your writing (customization)
The service is offered for R&D / non-diagnostic purposes only and results must not be utilized to inform patient management decisions. With our advanced sequencing platforms and years of industry experience, we guarantee you high-resolution data and comprehensive bioinformatics analysis. If you have additional requirements or questions, do not hesitate to contact us.

Table of HLA Type Details (Mayor NP et al., PLoS One. 2015)

Distribution of HLA Allelic Presentation (Nguyen A et al., Journal of virology, 2020)

Distribution of Allelic Presentation for All HLA Alleles and Individually for HLA-A, HLA-B, and HLA-C (Nguyen A et al., Journal of virology, 2020)
References
- Mayor NP, Robinson J, McWhinnie AJM, et al. HLA Typing for the Next Generation. PLoS One. 2015;10(5):e0127153. https://doi.org/10.1371/journal.pone.0127153
- Nguyen A, David JK, Maden SK, et al. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. Journal of virology. 2020 Jun 16;94(13):10-128. https://doi.org/10.1128/jvi.00510-20
1. What information can be obtained through HLA gene sequencing and information analysis?
Through NGS, HLA typing enables precise identification of specific allele information at each HLA locus, distinguishing between classical and non-classical HLA genes. It also determines specific combinations of HLA allele groups inherited together on a single chromosome. Additionally, NGS-based methods detect previously unreported alleles and ascertain whether each HLA locus is homozygous or heterozygous.
NGS-based HLA typing methods offer advantages of high accuracy, resolution, throughput, and cost-effectiveness. However, the accuracy of NGS-based HLA typing heavily depends on bioinformatics analysis methods, showing significant variability among different approaches.
2. What are the main methods of genotyping?
Serological typing: Focused on the specificity of HLA antigens, this method mainly employs HLA microcytotoxicity tests to analyze HLA types. Serological methods are vital for determining HLA typing and serve as the internationally recognized standard technique.
DNA typing: Concentrating on the analysis of genes themselves, this approach encompasses two main methods: those based on nucleic acid sequence identification and those based on sequence molecular configuration. The nucleic acid sequence identification methods primarily include PCR-RFLP, PCR-SSO, PCR-SSP, PCR-SBT, and the recently emerging next-generation sequencing methods.
3. Is NGS-based HLA typing suitable for clinical applications?
Indeed, NGS-based HLA typing is increasingly being integrated into clinical practice, particularly for organ transplantation, disease association studies, and pharmacogenomics. Its capability to provide high-resolution and precise allele determination enhances compatibility assessments and supports the development of individualized therapeutic strategies.
4. What level of precision is typically required in genotyping?
The specificity of the genotyping should ideally meet the criteria set by the recording of HLA types in the IMGT/HLA database. Differences in the four-digit typing correspond to variations in the encoded proteins, which generally fulfill the demands of scientific research. However, for studies necessitating higher accuracy or specific cases in transplant matching, a six-digit typing may be necessary to acquire more detailed allelic information.
High-Resolution HLA Typing of HLA-A, -B, -C, -DRB1, and -DQB1 in Kinh Vietnamese by Using Next-Generation Sequencing
Journal: Frontiers in genetics
Impact factor: 4.772
Published: 30 April 2020
Background
HLA genes, located on the short arm of chromosome 6, encode major histocompatibility complex proteins crucial for immune function. They are categorized into Class I (A, B, C) presenting intracellular antigens and Class II (DR, DQ, DP) presenting extracellular antigens to T cells. These genes exhibit high polymorphism and play key roles in immune-related diseases, tumor development, organ transplant outcomes, and drug reactions. Traditional DNA-based HLA typing methods include SSP, SSO, RFLP-PCR, and SBT, with NGS significantly enhancing resolution and accuracy. NGS overcomes phase ambiguity, enabling high-resolution typing of HLA alleles on a large scale. For the Southern Kinh Vietnamese population, high-resolution 3-field NGS typing of HLA-A, -B, -C, -DRB1, and -DQB1 is crucial to determine allele and haplotype frequencies accurately.
Materials & Methods
Sample Preparation
- 101 unrelated healthy individuals
- Venous blood
- Peripheral blood leukocytes
- DNA extraction
Sequencing
- Library preparation
- Next-generation sequencing
- HLA typing
- Single-locus analysis
- Multiple-locus analysis
- Principal component analysis
- Statistical analysis
Results
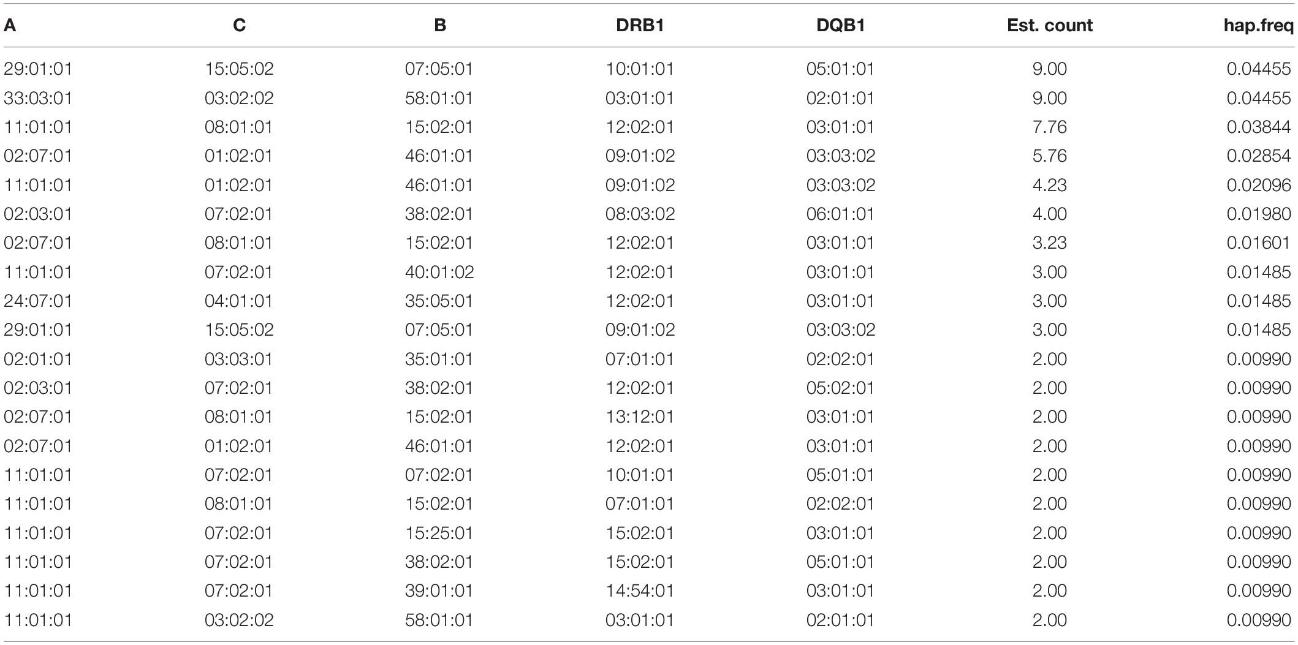
The study identified 28 HLA-A, 41 HLA-B, 21 HLA-C, 26 HLA-DRB1, and 25 HLA-DQB1 alleles. Among HLA-A alleles, A∗11:01:01, A∗24:02:01, and A∗33:03:01 were most frequent. For HLA-B, B∗15:02:01, B∗46:01:01, B∗58:01:01, B∗40:01:02, B∗38:02:01, and B∗07:05:01 were predominant. The top HLA-C alleles included C∗07:02:01, C∗01:02:01, and C∗08:01:01. HLA-DRB1∗12:02:01 and HLA-DQB1∗03:01:01 were highly prevalent. All loci tested exhibited no significant departure from Hardy–Weinberg equilibrium (p > 0.05). Neutrality tests also showed no significant results across the loci (p > 0.05).
Table 1. HLA frequency in the Kinh population (n = 101) (AF: allele frequency).
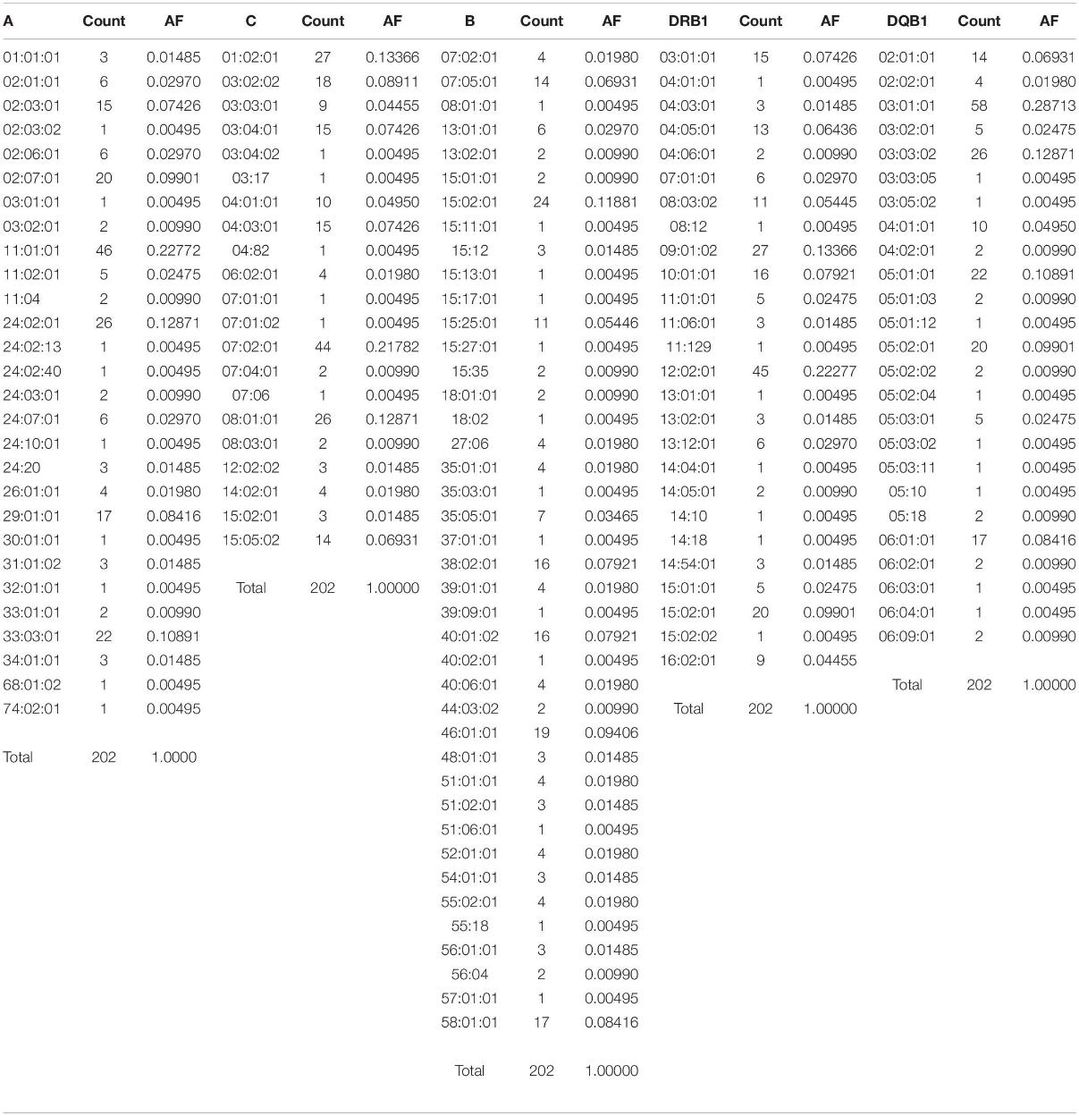
Table 2. Results of the Ewens–Watterson homozygosity test of neutrality.
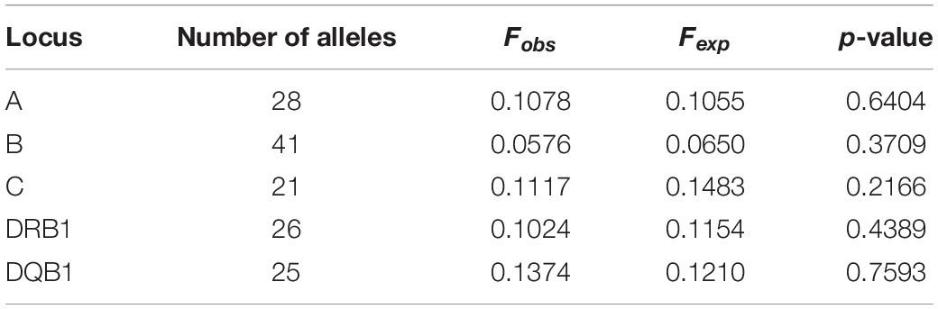
The study identified common haplotypes of HLA alleles across two-, three-, and five-locus combinations. Significant associations were found for all combinations tested (p < 0.001), highlighting strong linkage disequilibrium.
Table 3. Haplotype frequencies of two-locus HLA.
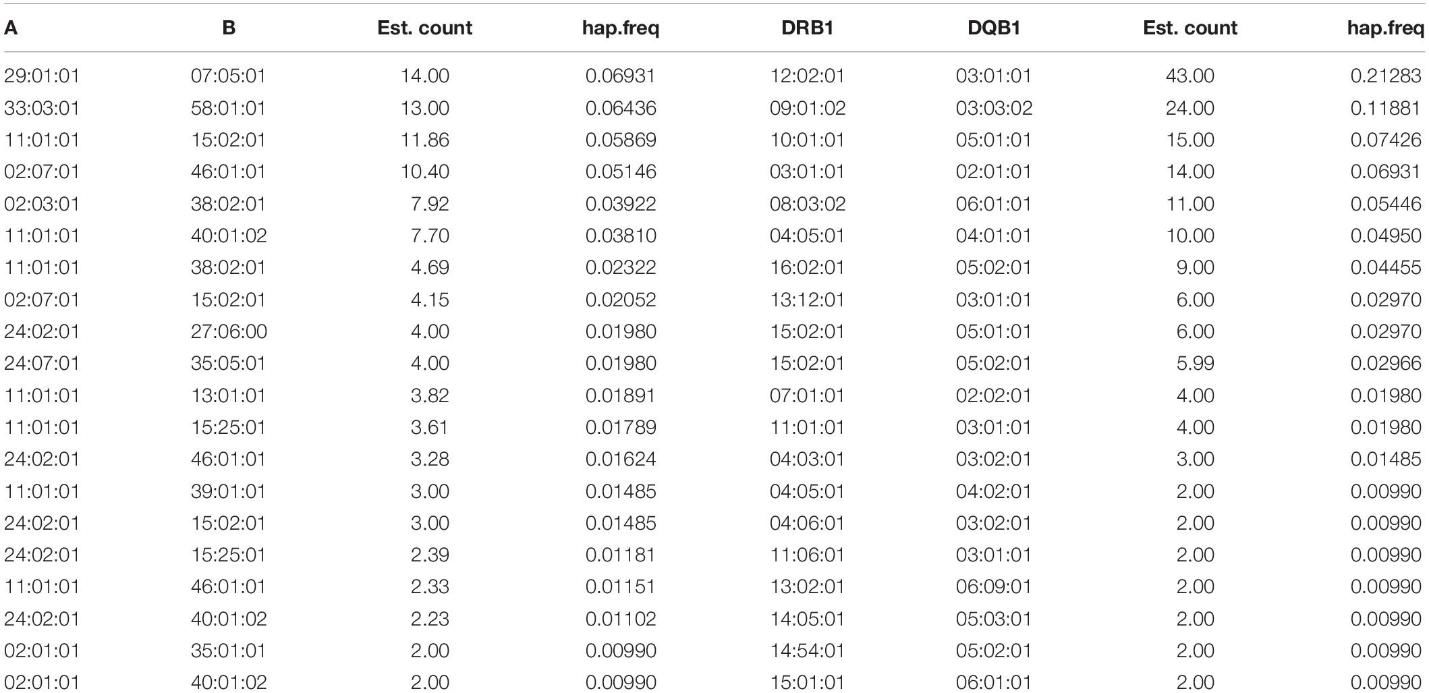
Table 4. Haplotype frequencies of three-locus HLA.
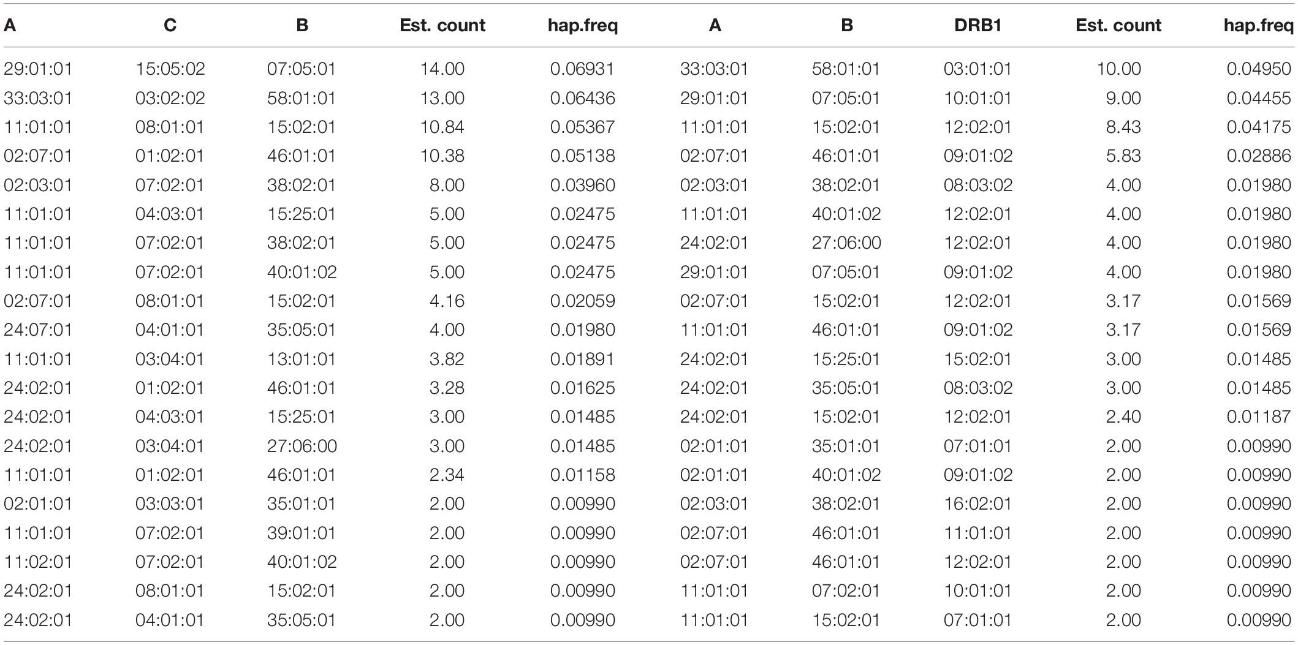
Table 5. Haplotype frequencies of five-locus HLA.
The study analyzed population genetics using LD estimates and PCA. LD analysis indicated strong associations within HLA classes, especially between C & B and DRB1 & DQB1 loci. PCA showed distinct genetic patterns among Asian populations, distinguishing South-East Asian, Han Chinese, Japanese, and South Korean groups based on HLA allele frequencies. Similar allele distributions were observed in northern and southern Kinh Vietnamese, mirroring patterns found in Japanese and South Korean populations.
Table 6. Pairwise linkage disequilibrium estimates.
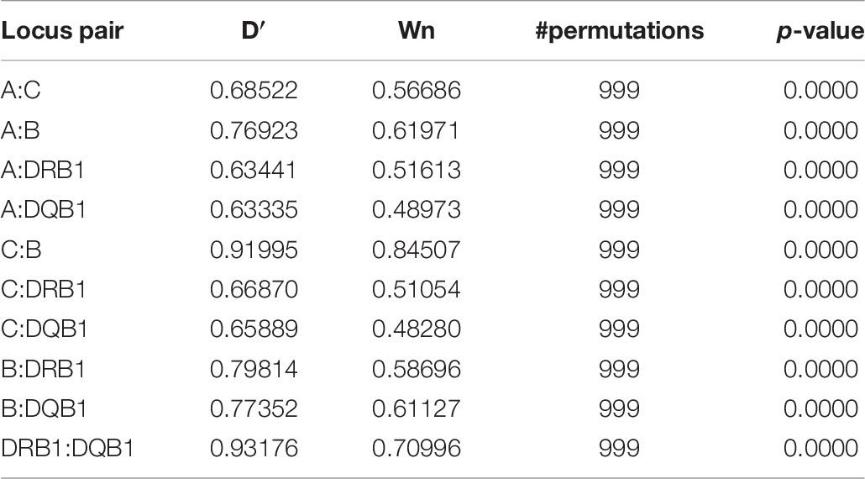
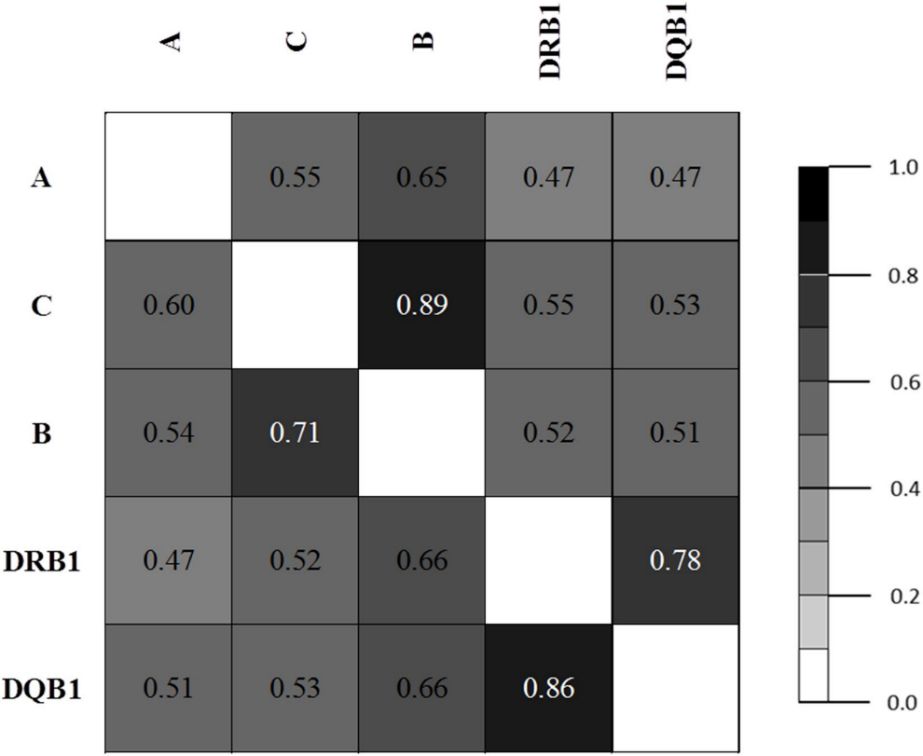 Figure 1. LD plot based on asymmetric linkage disequilibrium (ALD) measures for HLA genes.
Figure 1. LD plot based on asymmetric linkage disequilibrium (ALD) measures for HLA genes.
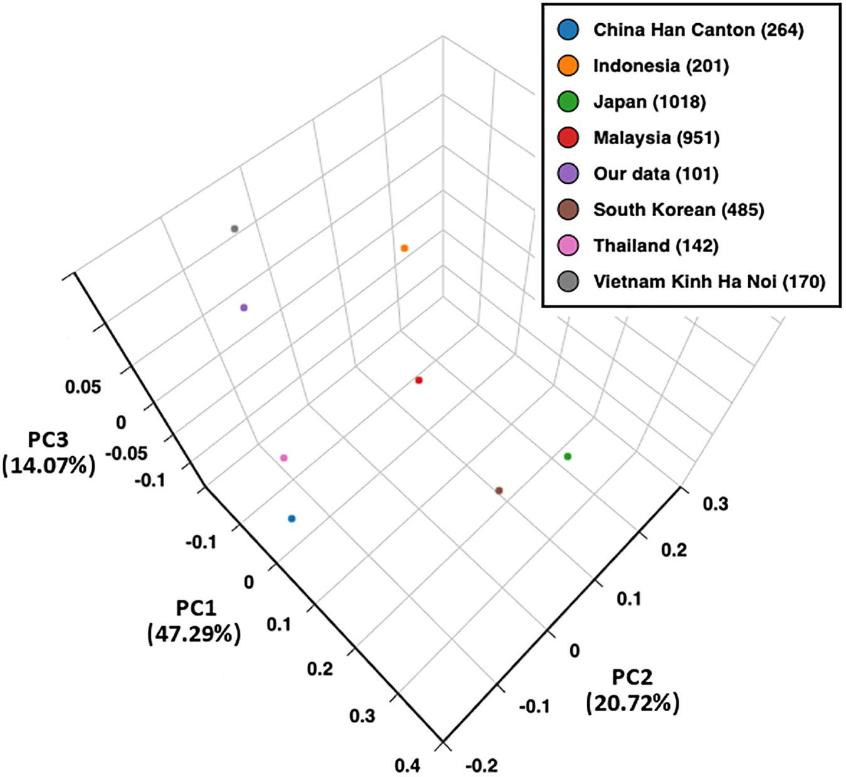 Figure 2. Principal component analysis (PCA) plot of eight populations based on HLA-A, -B, and -DRB1 allele frequencies.
Figure 2. Principal component analysis (PCA) plot of eight populations based on HLA-A, -B, and -DRB1 allele frequencies.
Conclusion
This study presents the first detailed analysis of HLA-A, -B, -C, -DRB1, and -DQB1 alleles and haplotypes in southern Kinh Vietnamese. It shows consistent HLA distributions across northern and southern Kinh populations in Vietnam, with unique features compared to Thai, Malaysian, and Indonesian populations. These findings are crucial for anthropology, immune diseases, transplantation, and drug reactions.
Reference
- Do MD, Le LG, Nguyen VT, et al. High-resolution HLA typing of HLA-A, -B, -C, -DRB1, and-DQB1 in Kinh Vietnamese by using next-generation sequencing. Frontiers in genetics. 2020, 11:383.
Here are some publications that have been successfully published using our services or other related services:
The HLA class I immunopeptidomes of AAV capsid proteins
Journal: Frontiers in Immunology
Year: 2023
Isolation and characterization of new human carrier peptides from two important vaccine immunogens
Journal: Vaccine
Year: 2020
Change in Weight, BMI, and Body Composition in a Population-Based Intervention Versus Genetic-Based Intervention: The NOW Trial
Journal: Obesity
Year: 2020
Sarecycline inhibits protein translation in Cutibacterium acnes 70S ribosome using a two-site mechanism
Journal: Nucleic Acids Research
Year: 2023
Identification of a Gut Commensal That Compromises the Blood Pressure-Lowering Effect of Ester Angiotensin-Converting Enzyme Inhibitors
Journal: Hypertension
Year: 2022
A Splice Variant in SLC16A8 Gene Leads to Lactate Transport Deficit in Human iPS Cell-Derived Retinal Pigment Epithelial Cells
Journal: Cells
Year: 2021
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
