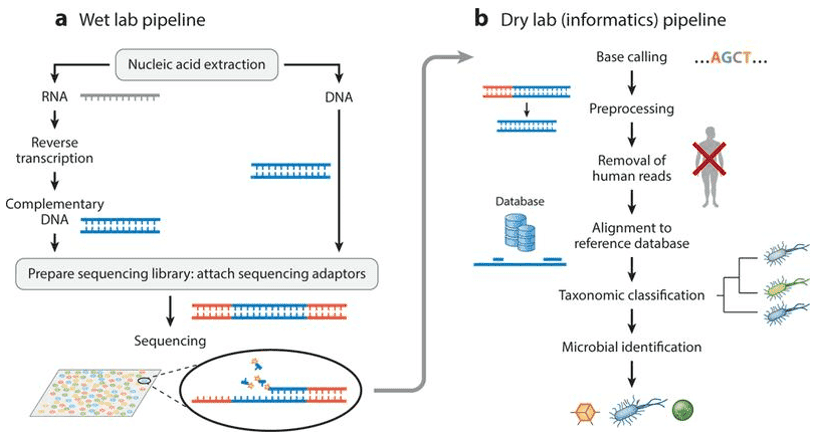
With the advent of research involving the human microbiome using high throughput sequencing of genomic DNA, there have been new discoveries that have given even more significance to human health in relation to the existing microbial flora in human bodies. In turn, there have been innovative therapeutic approaches to diseases based on the status and composition of the human microbiome. These discoveries are aided by studies that use DNA sequencing technologies like 16S rRNA sequencing, also known as 'amplicon-based' or 'targeted', and metagenomic whole genome shotgun (WGS) sequencing, both of which identify and characterize the different microbial communities within the human bodies.
The first sequencing method mentioned, referred to as 'targeted or amplicon-based sequencing', utilizes the genome from the sample and searches for the 16S rRNA sequence within it with sequence reads similar to that of the target bacterial taxa. It is used mainly to characterize the diverse and layered microbial communities in varying sampling sites in the human body. The analysis is limited though, as it is less precise at the species level. Another method is called 'metagenomic whole genome shotgun sequencing', where metagenome refers to the whole genomic DNA of the microbial communities present in the environment. It mainly functions to elucidate the varying functions and pathways of the human microbiome, in order to further understand how the existing microbial communities in humans affect human health. Together, both these high throughput sequencing techniques have unveiled some of the mysteries surrounding the human microbiome, also referred to as the 'second genome', thus, surpassing the limitations of culture-dependent techniques to identify and characterize microorganisms.
Metagenomic studies have postulated that there is significant coordination between the gut microbiome and the development of human diseases, and the involvement of the gut microbiome in moderating basic homeostatic processes, immunity, and signaling patterns within the body. As such, dysbiosis, or a malfunction in the microbiome, is also often observed to be tied to different human diseases. More specifically, studies on the microbiome found in the human gut have observed to have a strong correlation to the food, dietary supplements, and medicine consumed by humans. In fact, the gut microbiome composition is altered or has an effect on the materials that humans ingest as these must first pass through the gut before it reaches its intended destination (also referred to as the first-pass metabolism).
Dietary additives, like the use of non-caloric artificial sweeteners (NAS), for example, may have adverse effects when encountering the gut microbiome. For instance, germ-free mice inoculated with gut microorganisms from the gut of mice fed regularly with NAS, or fecal microorganisms exposed to NAS outside of a living organism, have a dysfunctional glucose metabolic pathway. In research studies involving humans, however, this effect was observed in only a portion of the subjects, as such, more studies need to be conducted to fully elucidate the effect of NAS on the gut microbiota. Another example of a food additive is emulsifiers, used in processed food to prolong shelf-life without sacrificing the physical integrity of the food product. When intake is excess as exhibited in mice, can cause physiological effects such as the thinning of the mucus-lining in the intestinal walls, and a change in the composition of the gut microbiota. This change in gut microbiome was seen to be simultaneous with other physical symptoms, some of which lead to heart problems. These symptoms were also replicated in animals that received microorganisms from the microbiome of animals that had been fed with emulsifiers, even if the recipient had not been treated with emulsifiers. This indicates that the gut microbiome may cause physiological symptoms like inflammation and those related to heart disease.
References
- Malla MA, Dubey A, Kumar A, et al. Exploring the human microbiome: The potential future role of next-generation sequencing in disease diagnosis and treatment. Frontiers in Immunology. 2019, 9:2868.