The human microbiome is the complete range of microorganisms (the microbiota) that live on and in humans. Specifically, it is the collection of microbial genomes that contribute to a human being's broader genetic picture, or metagenome. Genomes that make up the human microbiome include archaea, bacteria, fungi, protozoa and non-living viruses.
Importance of Human Microbiome
Humans do not have all the enzymes to digest the food thus the microbes in the gut break down many of the proteins, lipids and carbohydrates into nutrients to be able to be absorbed. The microbes also generate beneficial compounds, like vitamins and anti-inflammatories that genomes cannot create. Fecal microbiota transplantation (FMT or fecal transplantation) is one of the common microbiome therapies that restores healthy bacteria in the colon by introducing stool from a healthy human donor through colonoscopy or enema.
Human Microbiome of Different Organs
Gut Microbiome: The most studied human microbiome is the gut biome due to the vast number of microorganisms present in the gastrointestinal tract (GIT). Two bacterial phyla, gram-negative Bacteroidetes and gram-positive Firmicutes, belong to the majority of bacteria found in the gut in adults. Other organisms present that are represented at subdominant levels include Actinobacteria, Fusobacteria, and Verrucomicrobia phylum.
Vagina Microbiome: Studies have shown that the vagina has more than 200 phylotypes and that Firmicutes, Bacteroidetes, Actinobacteria, and Fusobacteria are the most predominant phyla.
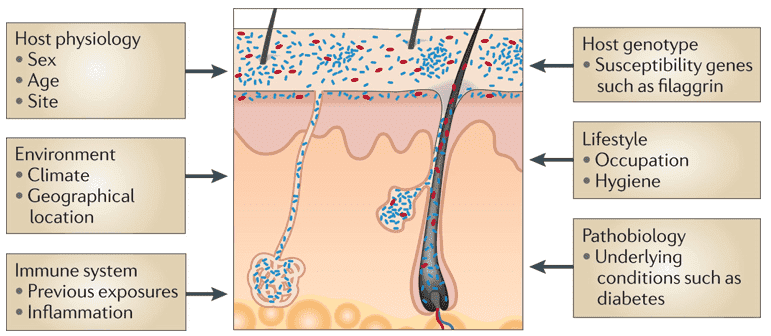
Skin Microbiome: The skin is the largest organ in the human body, colonized by even more than 100 microbial phylotypes, most of which are harmless or even advantageous to their host. In relation to skin color, race, and geographic location, phylotypes, microbial abundance, colonization and microbe diversity differ. Gram-positive Staphylococcus epidermidis is predominant on human epithelia while Propionibacterium acnes is predominant in sebaceous follicles.
Placenta Microbiome: The fetus and the intrauterine environment have historically been considered sterile. Even so, the first microbe profile in sustainable-term pregnancy recognized a distinct microbiome niche in normal placenta. It is composed of non-pathogenic commensal microbiota from Firmicutes, Tenericutes, Proteobacteria, Bacteroidetes and Fusobacteria phyla.
Oral Microbiome: The human oral cavity consists of various habitats, such as teeth, gingival sulcus, tongue, cheeks, hard and soft palate, and tonsils are colonized by bacteria. Some of the taxa present in human oral microbiome are the following: Actinobacteria, Bacteroidetes, Chlamydiae, Chloroflexi, Euryarchaeota, Firmicutes, Fusobacteria, Proteobacteria, Spirochaetes, SR1, Synergistetes, and Tenericutes.
Lung Microbiome: The lung is a warm environment that is exposed each day to 7000 liters of various microbe-ridden air. Using culture-independent techniques, healthy lungs are said to have predominant phyla Bacteroidetes and Firmicutes.
Research Methods Used in Exploring Human Microbiome
Metagenomic Sequencing: The biotechnological approach to the study of genomic sequences of uncultivated microbes directly from their natural sources is Metagenomics. The approach to metagenomics has the potential to uncover completely new genes, gene families, and encoded proteins that could be of biotechnological and pharmaceutical importance. Potential medical uses include the diagnosis of contagious diseases, monitoring of outbreak and infection control, discovery of possible mutation, and infection.
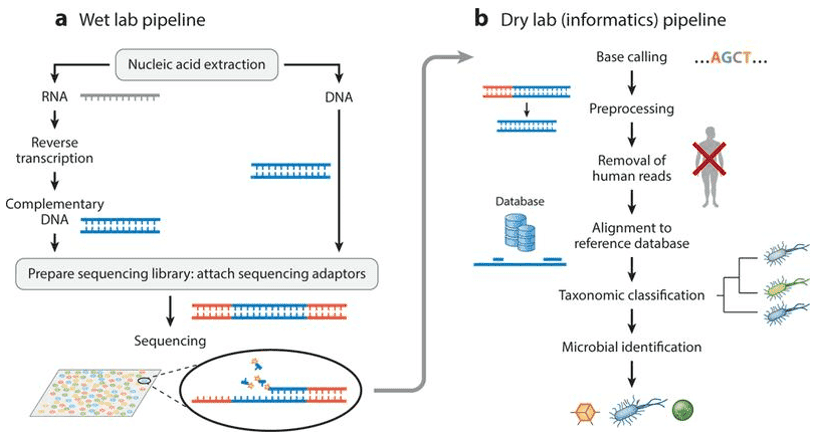 Figure 1. Schematic of the generalized workflow of metagenomic next-generation sequencing for diagnostic clinical use (Pathol, 2019)
Figure 1. Schematic of the generalized workflow of metagenomic next-generation sequencing for diagnostic clinical use (Pathol, 2019)
Metatranscriptomics: The metatranscriptome provides a snapshot of the gene expression in a given sample at a given moment and under specific conditions by capturing the total mRNA. It provides an informative perspective since it has the capability to provide details regarding populations that are as it can reveal details of populations. Symbiotic bacteria (normal flora) play a crucial role in protecting us from pathogens, but some harmful microorganisms can overcome protective host responses and trigger pathological effects under certain conditions. Microbial population analysis using metatranscriptomics can be used as an indicator of individual health status and as an instrument for the preventing, diagnosing, and treating certain diseases.
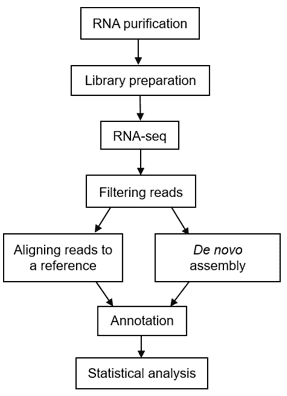 Figure 2. A typical workflow of metatranscriptomic sequencing (Peimbert M, et al. 2016)
Figure 2. A typical workflow of metatranscriptomic sequencing (Peimbert M, et al. 2016)
References
- Kho, Z. Y., Lal, S. K. The Human Gut Microbiome – A Potential Controller of Wellness and Disease. Frontiers in Microbiology. 2018.
- Manuzak JA, Hensley-McBain T, Zevin AS, et al. Enhancement of Microbiota in Healthy Macaques Results in Beneficial Modulation of Mucosal and Systemic Immune Function. J Immunol. 2016.
- Turnbaugh, P. J., Ley, R. E., Hamady, M., Fraser-Liggett, C. M., et al.. The Human Microbiome Project. Nature. (2007), 449(7164), 804–810.


 Figure 1. Schematic of the generalized workflow of metagenomic next-generation sequencing for diagnostic clinical use (Pathol, 2019)
Figure 1. Schematic of the generalized workflow of metagenomic next-generation sequencing for diagnostic clinical use (Pathol, 2019) Figure 2. A typical workflow of metatranscriptomic sequencing (Peimbert M, et al. 2016)
Figure 2. A typical workflow of metatranscriptomic sequencing (Peimbert M, et al. 2016)