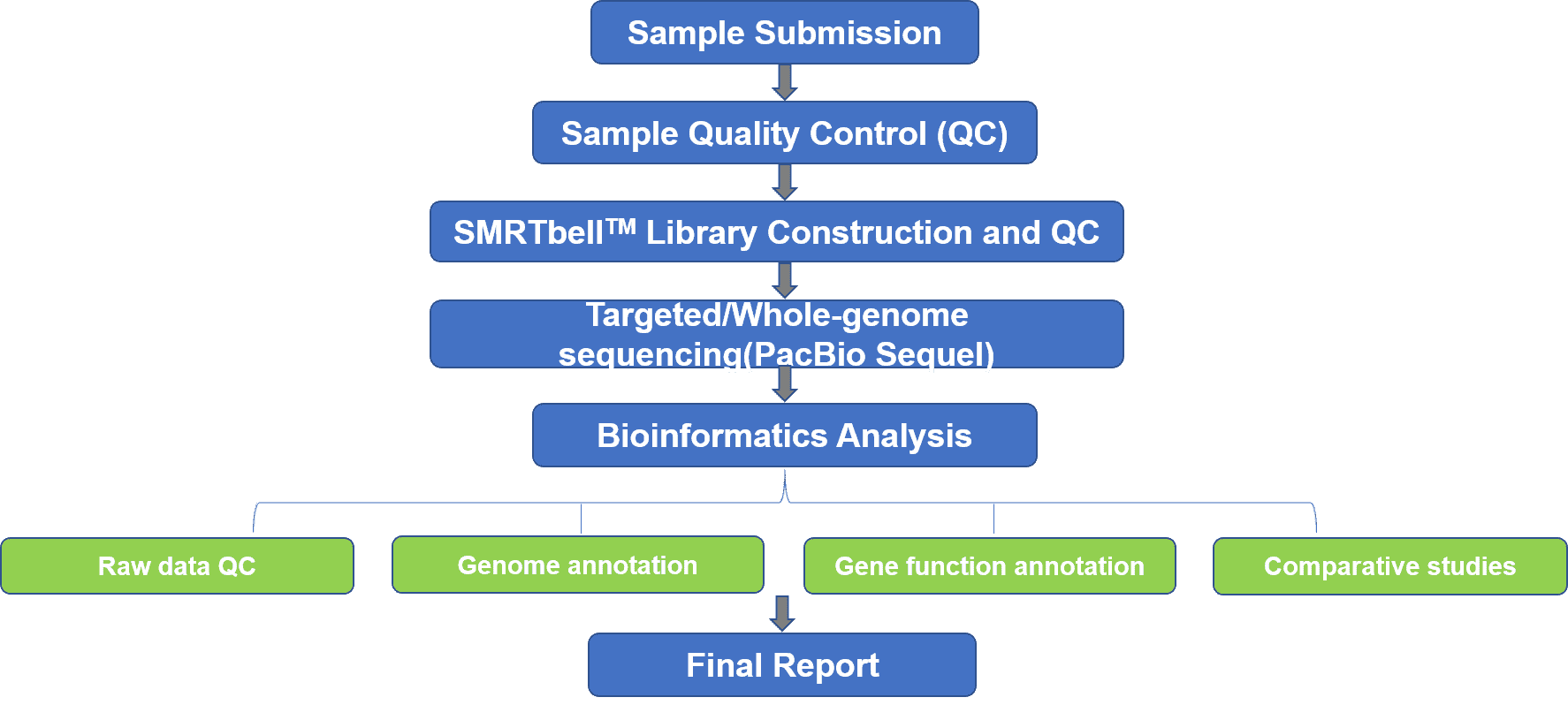
The microbial genome is smaller than that of eukaryotes. Most bacteria have only one circular chromosome, with the exceptions of other bacteria with more than one (several linear chromosomes or combinations of circular and linear chromosomes). The presence of plasmids (the circular extra-chromosomal DNA and the bacterial genome) is another major characteristic of prokaryotic genomes, which mediating the rapid evolution of microorganisms and can be transferred by horizontal DNA transfer. Our SMRT-based genome sequencing, using technology from PacBio Biosciences, can provide accurate contiguous sequences for complete bacterial, viral, and fungal DNA or RNA genomes. The PacBio SMRT sequencing technology is characterized by unparalleled raw read accuracy, read length and read depth, which is helpful for the reconstruction of high-quality drafts and complete microbial genomes.
Based on the PacBio SMRT platform, we provide microbial targeted and whole-genome sequencing services to analyze challenging genomic regions like structural variants and repeat sequences for high-quality assemblies, perform accurate microbial identification, clarify the role of phage insertion, transposon insertion and other structural variants in the evolution of virulence, and recover plasmids to track transmission paths and drug resistance. Our SMRT-based genome analysis can be used to redefine the genetic and biochemical diversity in complex communities, elucidate virus-host interactions, uncover new taxonomic groups and metabolic capabilities, and determine how functions are distributed across environmental gradients.