Escherichia coli, a well-known model organism in microbiological research, gained prominence following the elucidation of the lactose operon model in 1962, a topic of particular interest that will be discussed comprehensively in the third and fourth sections of this manuscript. Beyond this model, the E. coli genome encompasses numerous other genes, unveiling the profound and intricate nature of genetic mysteries.
As a subject of scientific inquiry, the E. coli genomic structure manifests as a closed circular chromosome with an approximate length of five million base pairs (5 Mbp) and encodes approximately 5,000 genes. This genomic configuration is notably compact, with approximately 90-95% of the DNA sequence actively involved in coding, thereby exhibiting scant intergenic sequences. This high-density gene arrangement highlights frequent utilization of most genes during transcription and translation processes, reflecting the optimized nature of the E. coli genome.
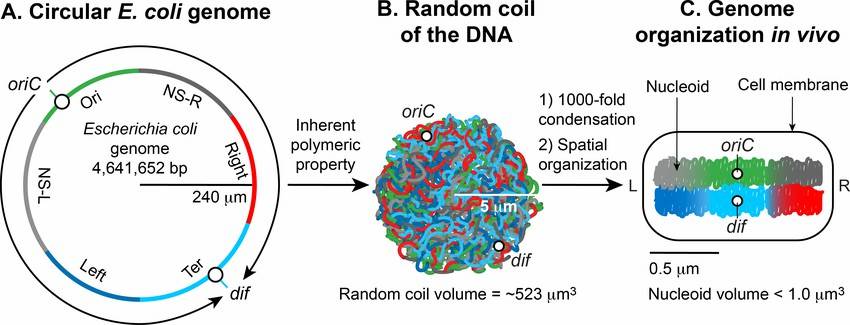 Formation of the Escherichia coli nucleoid A. (Subhash C. Verma et al,. 2019)
Formation of the Escherichia coli nucleoid A. (Subhash C. Verma et al,. 2019)
Structural Characteristics of Prokaryotic Genes
Linear Gene Architecture
In prokaryotic organisms, genes typically manifest as continuous linear sequences devoid of introns, in stark contrast to the segmented gene architecture prevalent in eukaryotes. The absence of introns obviates the need for extensive splicing of transcriptional products, thereby facilitating the direct engagement of these transcripts in the translation process, which markedly enhances the efficiency of gene expression.
Additionally, prokaryotic genomes characteristically contain relatively short intergenic non-coding regions, with coding regions being densely packed. This arrangement results in a highly compact gene alignment.
Operon Structure
Prokaryotic genes are frequently organized in clusters, forming structures known as “operons,” wherein multiple functionally related genes share a common promoter and regulatory sequences, permitting their concurrent transcription into a single polycistronic mRNA. For instance, the lactose operon (lac operon) encompasses genes such as lacZ, lacY, and lacA, which are co-transcribed on the same mRNA strand. This organization enables cellular systems to coordinate the expression of functionally analogous proteins effectively.
The operon structure endows prokaryotic cells with the capacity for rapid environmental adaptation, thereby demonstrating efficient metabolic adaptability.
1. Structure of the Lactose Operon
The lactose operon consists of several key components: three structural genes—lacZ, lacY, and lacA—a promoter (lacP), an operator (lacO), and a regulatory gene, lacI. The lacI gene is crucial because it codes for the lactose repressor protein, LacI. This protein can bind to the operator site, essentially blocking RNA polymerase from attaching and thus halting the transcription of the genes responsible for lactose metabolism. Without this regulation, cells might waste energy expressing these genes when lactose isn't available.
2. Regulation Mechanism of the Lactose Operon
When lactose is present, it's broken down into galactose and glucose, but that's not all. In the process, allolactose is also produced, and this molecule plays a crucial role. Allolactose acts as an inducer, binding to the LacI repressor protein. This binding triggers a structural change in LacI, causing it to detach from the operator sequence. With LacI no longer blocking the operator, RNA polymerase can freely bind and begin transcribing the genes lacZ, lacY, and lacA—genes that are all involved in the metabolism of lactose. This elegant mechanism ensures that the cell only ramps up the machinery for lactose digestion when it's actually needed.
3. Role of cAMP and CRP in the Lactose Operon
Cyclic adenosine monophosphate (cAMP) and its partner, cAMP receptor protein (CRP), work together to fine-tune the expression of the lactose operon. When glucose levels are low, cAMP levels rise, and this spike enables the formation of the CRP-cAMP complex. This complex, in turn, activates the expression of the lactose operon, making the machinery for lactose metabolism ready to go. But when glucose is plentiful, the opposite happens: cAMP levels drop, leading to a decrease in the CRP-cAMP complex's activity. As a result, the expression of the lactose operon slows down, preventing unnecessary use of resources when glucose is available as an energy source. This delicate balance ensures the cell adapts efficiently to the available nutrients.
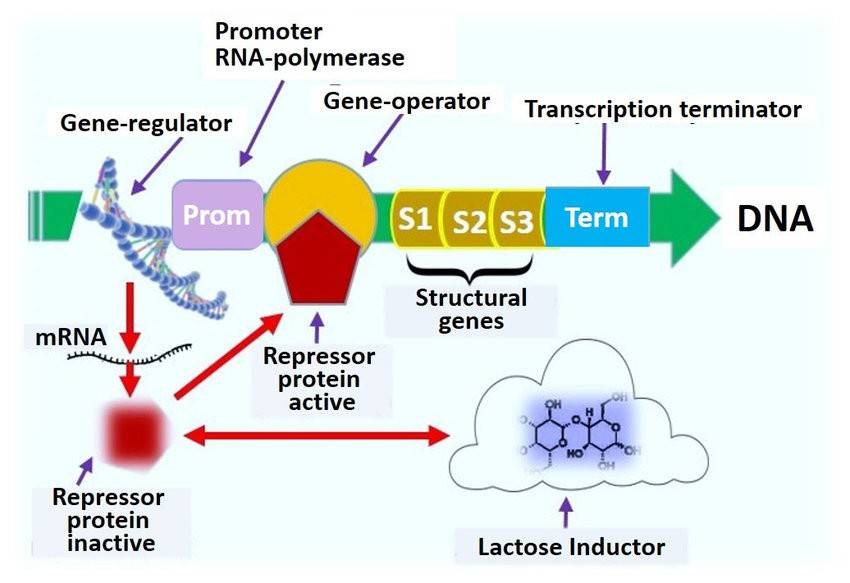 Functional diagram of the lactose lac operon of Escherichia coli in microbial biosensors. Andryukov et al,. 2020)
Functional diagram of the lactose lac operon of Escherichia coli in microbial biosensors. Andryukov et al,. 2020)
Regulatory Region
In prokaryotes, gene regulation often happens in regions located just upstream of the gene, where you'll find both the promoter and operator elements. These regulatory regions are typically short, with key sites near the -10 and -35 positions of the promoter, which are essential for RNA polymerase to bind. The sequence of the promoter plays a crucial role—it essentially sets the stage for how strongly RNA polymerase will attach, directly impacting gene expression levels. Operators, on the other hand, are like docking stations for regulatory proteins. These can be repressors or activators, and their job is to either enhance or block transcription by binding to or releasing from the operator. This operon structure gives prokaryotic cells an impressive ability to adapt quickly to their environment, optimizing their metabolism for efficiency when needed most.
Promoters and Terminators
The promoter region is the critical docking site for RNA polymerase, and it contains two important elements: the -10 box, also known as the Pribnow box, with its TATAAT sequence, and the -35 box with the sequence TTGACA. These sequences act as the signals that kick-start transcription. Meanwhile, at the other end of the gene, the 3' region harbors sequences responsible for terminating transcription. Prokaryotes have two main ways to stop transcription: intrinsic termination and Rho-dependent termination. Intrinsic terminators feature a G-C rich hairpin loop, which essentially causes RNA polymerase to release. On the other hand, Rho-dependent termination relies on the Rho factor, a protein that binds to the RNA and helps it detach from the polymerase, wrapping up transcription.
Plasmid DNA
Aside from chromosomal genes, many prokaryotic organisms, notably bacteria, possess additional circular DNA molecules known as plasmids. Plasmids carry genes that, while not essential, provide an adaptive advantage such as antibiotic resistance genes and virulence factors. Plasmids can be transferred between cells through horizontal gene transfer, enhancing bacterial adaptability.
Lack of Complex Modifications
The gene expression process in prokaryotic organisms generally bypasses the requirement for mRNA processing and modifications, such as 5' capping or 3' polyadenylation. Consequently, RNA transcripts directly proceed to the translation phase, thereby further augmenting expression efficiency.
Unveiling the Complexity of Prokaryotic Genomes: Implications for Microbial Sequencing and Beyond
In wrapping up, the genomic architecture of prokaryotic organisms, exemplified by Escherichia coli, reveals just how finely tuned these organisms are to thrive in their environments. From the efficient gene arrangement to the delicate regulatory controls like operons, prokaryotic genomes are crafted for rapid adaptability and metabolic precision. The molecular choreography of gene expression—whether orchestrated by the lactose operon or through the interplay of regulatory proteins and small molecules—shows just how sophisticated bacterial control mechanisms are at the cellular level.
As we delve deeper into these genetic processes, microbial sequencing technology has emerged as a game-changer. The ability to decode microbial genomes with precision has transformed our approach to studying microbial diversity, uncovering resistance mechanisms, and even opening the door to new therapeutic possibilities. This leap in sequencing technology not only propels scientific discovery but also plays a pivotal role in clinical microbiology, where understanding pathogens' genetic makeup is vital for tackling infectious diseases. As these sequencing techniques continue to advance, we are on the cusp of unlocking even more intricate layers of microbial life—offering fresh insights that are set to impact both our understanding of biology and the future of human health.


 Formation of the Escherichia coli nucleoid A. (Subhash C. Verma et al,. 2019)
Formation of the Escherichia coli nucleoid A. (Subhash C. Verma et al,. 2019) Functional diagram of the lactose lac operon of Escherichia coli in microbial biosensors. Andryukov et al,. 2020)
Functional diagram of the lactose lac operon of Escherichia coli in microbial biosensors. Andryukov et al,. 2020)