Non-metric multidimensional scaling, or NMDS, is known to be an indirect gradient analysis which creates an ordination based on a dissimilarity or distance matrix. It attempts to represent the pairwise dissimilarity between objects in a low-dimensional space, unlike other methods that attempt to maximize the correspondence between objects in an ordination. NMDS is a rank-based approach which means that the original distance data is substituted with ranks. It is considered as a robust technique due to the following characteristics: (1) can tolerate missing pairwise distances, (2) can be applied to a dissimilarity matrix built with any dissimilarity measure, and (3) can be used in quantitative, semi-quantitative, qualitative, or even with mixed variables.
Application in Bioinformatics
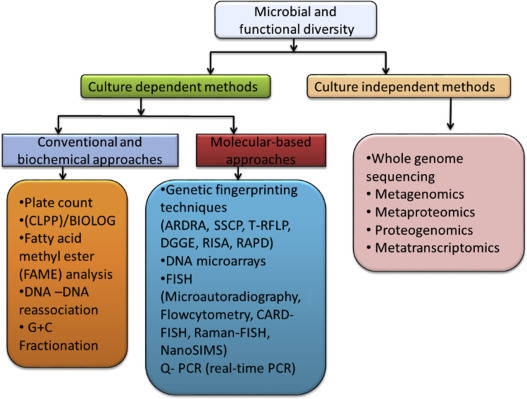
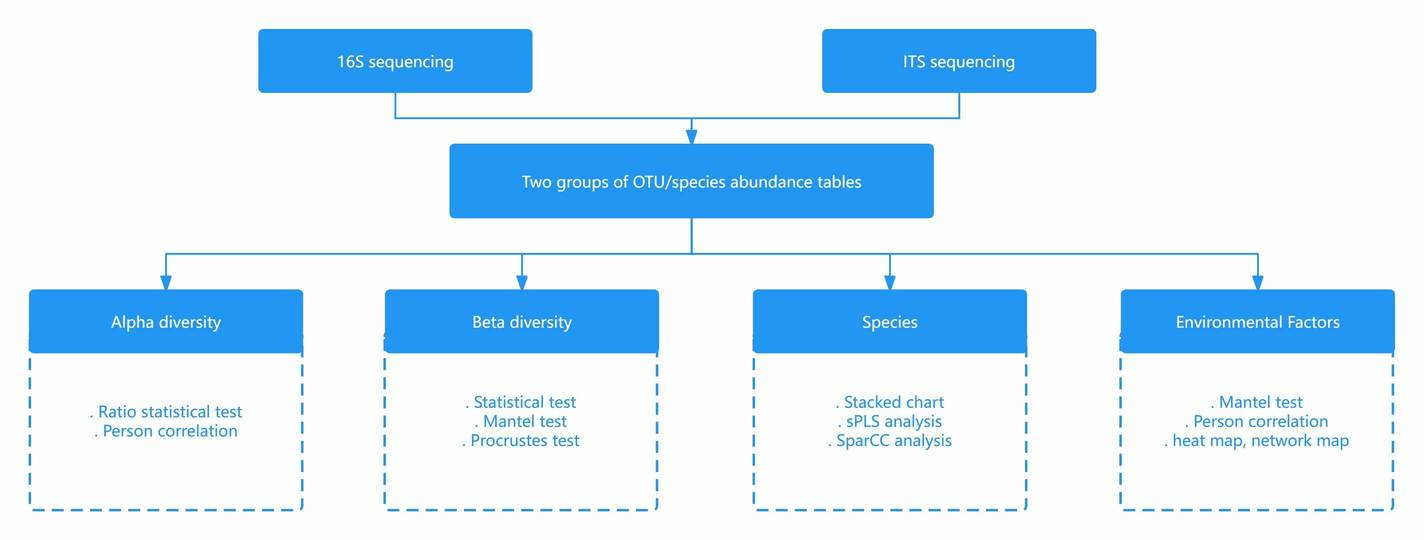
Several studies have revealed the use of non-metric multidimensional scaling in bioinformatics, in unraveling relational patterns among genes from time-series data. Specifically, the NMDS method is used in analyzing a large number of genes. Some studies have used NMDS in analyzing microbial communities specifically by constructing ordination plots of samples obtained through 16S rRNA gene sequencing.
Non-metric Multidimensional Scaling vs. Other Ordination Methods
In NMDS, there are no hidden axes of variation since a small number of axes are chosen prior to the analysis, and the data generated are fitted to those dimensions. Second, NMDS is a numerical technique that solves and stops computing when an acceptable solution has been found. This is different from most of the other ordination methods which results in a single unique solution since they are considered analytical. Third, NMDS ordinations can be inverted, rotated, or centered into any desired configuration since it is not an eigenvalue-eigenvector technique. Unlike correspondence analysis, NMDS does not ordinate data such that axis 1 and axis 2 explains the greatest amount of variance and the next greatest amount of variance, and so on, respectively. Lastly, NMDS makes few assumptions about the nature of data and allows the use of any distance measure of the samples which are the exact opposite of other ordination methods.
Limitations of Non-metric Multidimensional Scaling
NMDS has two known limitations which both can be made less relevant as computational power increases. First, it is slow, particularly for large data sets. Second, it can fail to find the best solution because it may stick on local minima since it is a numerical optimization technique. Although, increased computational speed allows NMDS ordinations on large data sets, as well as allows multiple ordinations to be run. This would greatly decrease the chance of being stuck on a local minimum.
Computation: The Kruskal's Stress Formula
Distances among the samples in NMDS are typically calculated using a Euclidean metric in the starting configuration. These calculated distances are regressed against the original distance matrix, as well as with the predicted ordination distances of each pair of samples. This was done using the regression method. This goodness of fit of the regression is then measured based on the sum of squared differences. That was between the ordination-based distances and the distance predicted by the regression. The most common way of calculating goodness of fit, known as stress, is using the Kruskal's Stress Formula: 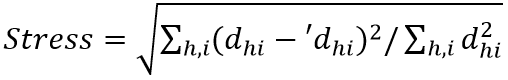 (where,dhi = ordinated distance between samples h and i; 'dhi = distance predicted from the regression)
(where,dhi = ordinated distance between samples h and i; 'dhi = distance predicted from the regression)
References
- Kim JS, Kim DS, Lee KC, et al. Microbial community structure and functional potential of lava-formed Gotjawal soils in Jeju, Korea. PloS one. 2018, 13(10):e0204761.
- Taguchi YH, Oono Y. Relational patterns of gene expression via non-metric multidimensional scaling analysis. Bioinformatics. 2015, 21(6):730-40.


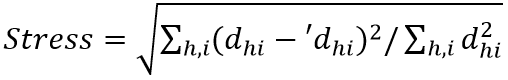 (where,dhi = ordinated distance between samples h and i; 'dhi = distance predicted from the regression)
(where,dhi = ordinated distance between samples h and i; 'dhi = distance predicted from the regression)