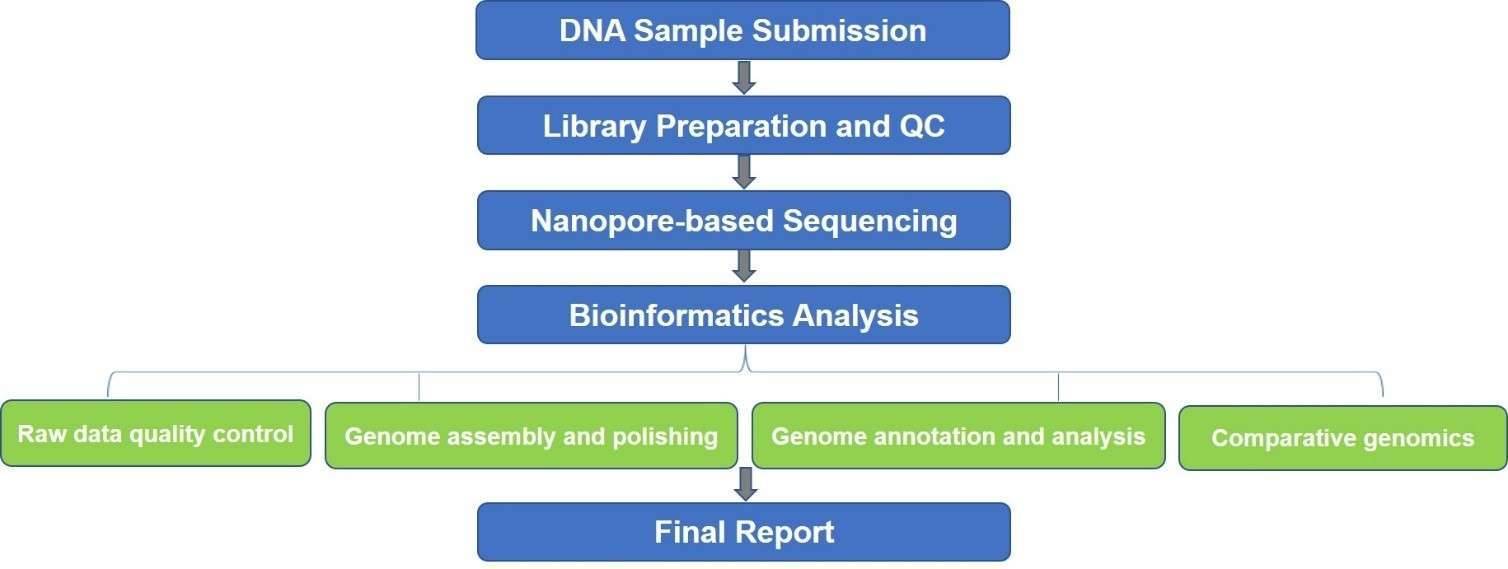
In recent years, there have been enormous achievements made in the DNA sequencing technologies, including the next-generation sequencing (NGS), 3rd-generation sequencing (also known as long-read sequencing), and the corresponding innovations in bioinformatics analysis. Those achievements are driving the development of an exciting field of biological research called the genome analysis. Oxford nanopore sequencing identifies DNA bases by measuring the current changes in electrical conductivity as nucleic acid molecules pass through a biological pore in membranes, generating much longer average reads (ultra-long reads, over 800,000 bases) than other DNA sequencing technologies. Our nanopore-based microbial genomics analysis can generate ultra-long reads (about 5.4 kb to 10 kb of average read length) that span challenging genomic regions. We also polish the microbial genome assembly with short reads generated by the Illumina system to improve accuracy to 99.96%.
Our nanopore-based microbial genome analysis services include microbial whole-genome sequencing and targeted sequencing, along with comprehensive bioinformatics tools to reconstruct bacterial, fungal, or viral genomes (de novo) and perform further genome-based analysis. The highly precise and complete genome assemblies enable the accurate microbial identification, genome structure evaluation, variants (phage insertion, transposon insertion, SVs, InDels, SNVs) identification along the evolution of virulence, understanding of the role of mobile elements in drug transmission and resistance, and gene function annotation. We are dedicated to helping you generate high-quality, closed-reference genomes, redefining the extent of genetic diversity and elucidating microbial genetics and evolution.