Navigation
- Home
- Services
- Microbial Diversity Analysis – 16S/18S/ITS Sequencing
- Metagenomics
- Microbial Whole Genome Sequencing
- Microbial Identification
- Microbial Characterization
- Microbial Functional Gene Analysis
- Microbial Epigenomics
- Antibiotic Resistance Genes (ARGs) Analysis Solution
- Microbial PacBio SMRT Sequencing
- Microbial Nanopore Sequencing
- Microbial Transcriptomics
- Microbial Bioinformatics
- Other
- Microbial Metabolomics Analysis Service
- Solutions
- Microecology and Human/Animal Health
- Environmental Microbiology Solutions
- Agricultural Microbiology Solutions
- Pharmaceutical Microbiology Solutions
- Industrial Microbiology Solutions
- Microecology and Cancer Research Solutions
- Microecology and Biofilms
- Pathogen Sequencing Solutions
- Environmental DNA (eDNA) Analysis Solution
- Products
- Resource
- Company
- Sample Submission Guidelines

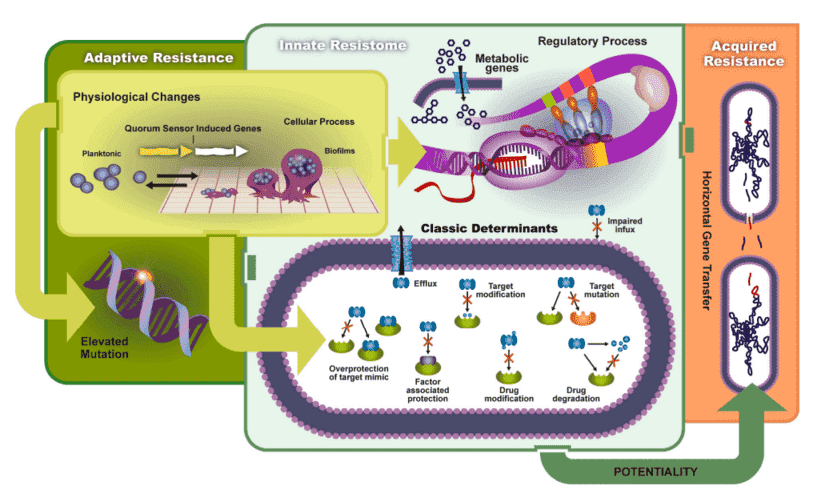 Figure 1. The transmission and mechanisms of antibiotic resistance and virulence can be divided into adaptive resistance, innate resistance, and acquired resistance. (Schroeder, 2017)
Figure 1. The transmission and mechanisms of antibiotic resistance and virulence can be divided into adaptive resistance, innate resistance, and acquired resistance. (Schroeder, 2017)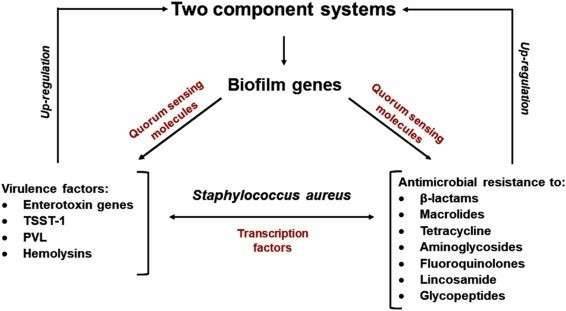 Figure 2. The relationship between virulence factors and antimicrobial resistance in Staphylococcus aureus. (Pérez, 2020)
Figure 2. The relationship between virulence factors and antimicrobial resistance in Staphylococcus aureus. (Pérez, 2020)