Antibiotic resistance represents a formidable challenge to global health, driven extensively by the selective pressure that enables bacteria to acquire antibiotic resistance genes (ARGs). In response to this pressing issue, CD Genomics offers advanced, genome-focused analysis services aimed at searching for antibiotic resistance genes. These rigorous services not only pinpoint existing ARGs but also play a pivotal role in uncovering novel ARGs, and in decoding the intricate molecular mechanisms that facilitate the transfer of these genes.
Our Advantages:
- Whole-genome sequencing and powerful bioinformatics analysis
- Multiple samples can be sequenced at a time with a streamlined workflow suitable for automation.
- State-of-the-art sequencing platforms are available like Illumina HiSeq/MiSeq, Nanopore MinION, and PacBio Sequel.
- Delivery of robust sequencing and data analysis results.
Introduction to Antibiotic Resistance Gene Screening
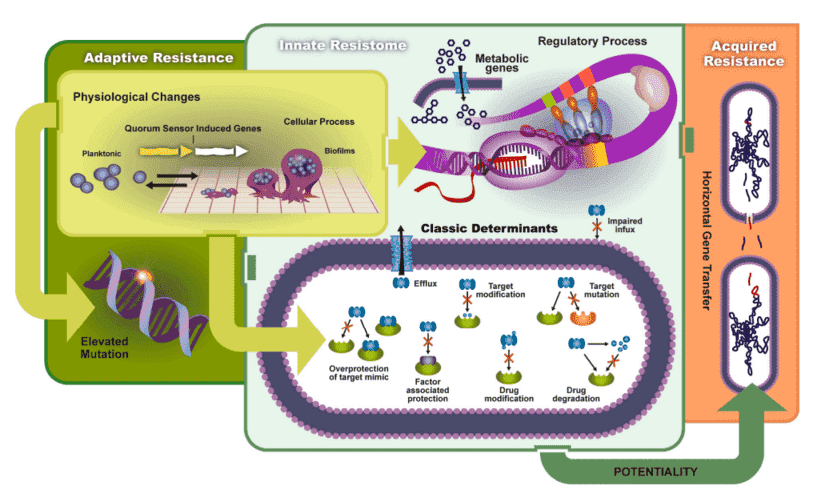
The relentless rise of antibiotic resistance (AR) poses a profound and growing threat to global public health, driven by the adaptive evolution of bacterial pathogens under the selective pressures of antimicrobial exposure. Understanding the molecular underpinnings of resistance is paramount, necessitating the accurate identification and thorough characterization of ARGs. These genetic determinants are pivotal in the ability of bacteria to withstand and proliferate in environments saturated with antibiotics.
ARGs are critical genetic components that can be inherited vertically through clonal expansion or acquired horizontally via mechanisms such as conjugation, transformation, or transduction. The rapid and widespread propagation of ARGs, often mediated by mobile genetic elements including plasmids and transposons, has exacerbated the global challenge of antibiotic resistance. This dissemination has rendered numerous conventional antimicrobial therapies increasingly ineffective, necessitating urgent and sophisticated approaches to detection and management.
At CD Genomics, we provide an extensive array of ARG screening services, leveraging state-of-the-art technologies such as quantitative PCR (qPCR) and next-generation sequencing (NGS), including techniques like metagenomic sequencing and whole genome sequencing. These methodologies facilitate the precise detection and in-depth characterization of ARGs, delivering critical insights that are integral to both academic research and clinical application, and ultimately, to the strategic control of antibiotic resistance.
How to Detect Antibiotic Resistance Genes
The detection of ARGs employs various methodologies that enhance the understanding and management of antibiotic resistance. Key techniques include:
1. Traditional Culture-Based Methods: These methods, such as antimicrobial susceptibility testing (AST), involve culturing bacteria to assess resistance. While informative, they require specialized labs and are limited to culturable organisms.
2. Molecular Techniques:
- Quantitative PCR (qPCR): qPCR quantitatively detects specific ARGs in samples, providing rapid and reliable results.
- Next-Generation Sequencing (NGS): NGS allows for high-throughput screening, enabling the identification and characterization of ARGs without the need for culturing, thus offering comprehensive insights into microbial resistance profiles.
3. Mass Spectrometry: Techniques such as MALDI-TOF-MS can identify bacterial species and their resistance profiles based on protein patterns, though they require accurate databases and pure cultures.
4. Emerging Techniques: Innovations like metagenomics, long-read sequencing, and WGS, along with mscRNA-seq technologies, enhance the detection of ARGs in complex samples, facilitating the study of resistance mechanisms in diverse microbial communities.
Applications of Antibiotic Resistance Genes Screening
The applications of antibiotic resistance genes screening include, but are not limited to, the following areas:
- Epidemiological Surveillance: ARG screening monitors resistance gene prevalence, aiding targeted public health interventions.
- Pharmaceutical Research: Understanding ARGs informs new antibiotic development and monitors resistance emergence in trials.
- Environmental Monitoring: Screening for ARGs in environmental samples assesses human impact on resistance gene spread and protects public health.
Partial results of our Antibiotic Resistance Genes Screening Sequencing service are shown below:


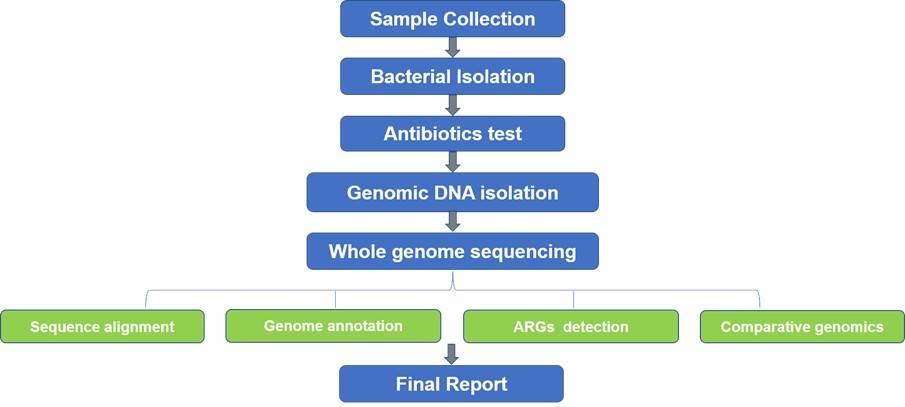

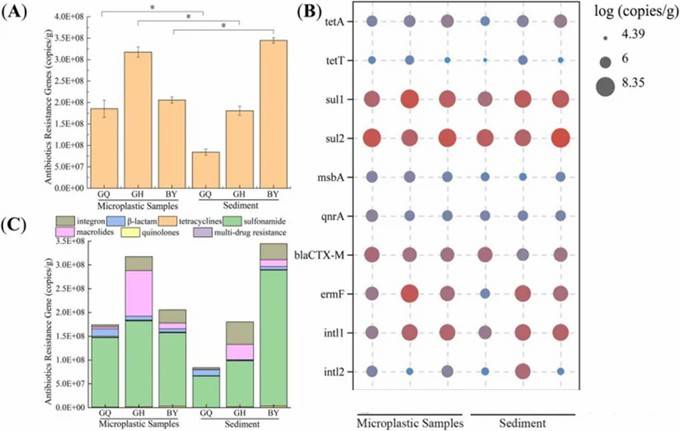 Fig 1. Spatial distribution of ARGs on MPs in different surrounding environments of mangroves.
Fig 1. Spatial distribution of ARGs on MPs in different surrounding environments of mangroves.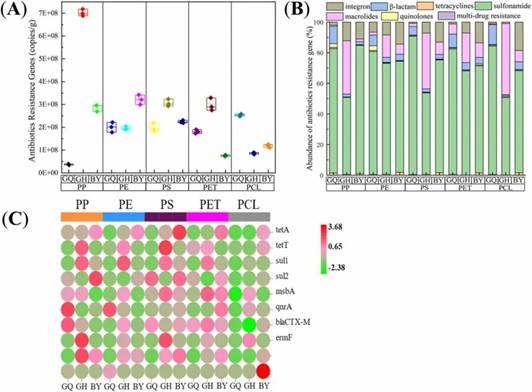 Fig 2. Distribution characteristics of ARGs on the surfaces of five different MPs.
Fig 2. Distribution characteristics of ARGs on the surfaces of five different MPs.