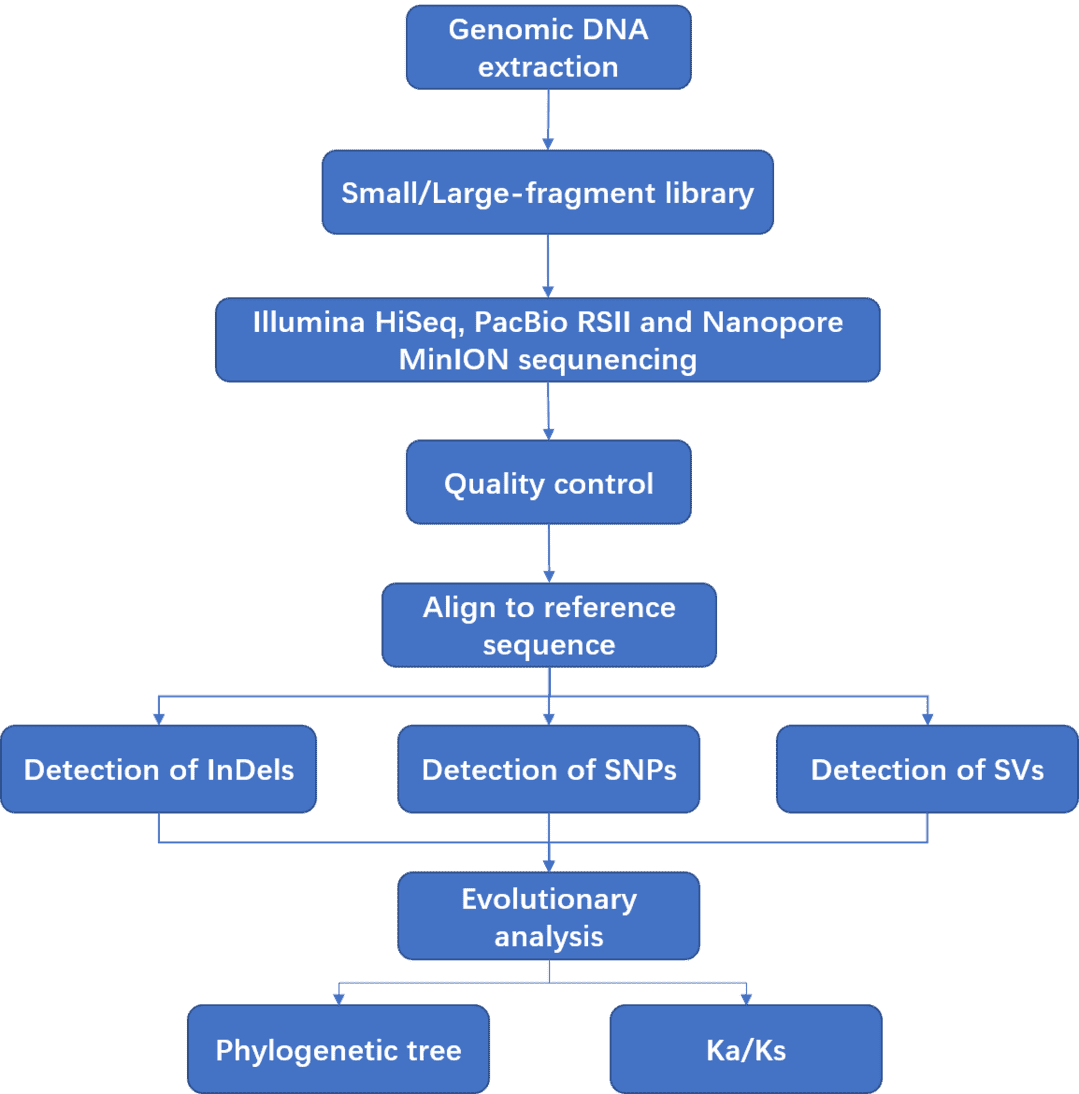
We provide fungal whole genome resequencing using a whole-genome shotgun strategy by next generation sequencing (Illumina HiSeq PE150/251, 100X) or long-read sequencing platforms (PacBio RSII or Nanopore MinION instrument). A large number of reads generated from sequencers are subjected to quality control, and the clean data are subsequently aligned to reference genome sequences. With powerful bioinformatics analysis, we can detect large variations including SNPs, InDels, SVs, CNVs, etc. Fungal whole genome resequencing is an effective and accurate, method to quickly sequence and analyze fungal genomes.
With the availability of more high-quality reference genome sequences from different species, the use of fungal genome resequencing becomes common. Fungal whole genome resequencing can quickly and efficiently detect both mutations and structural variations on a global genome. Comparative genomic analysis can identify differentially expressed genes through gene ontology analysis and explore the evolutionary relationships and biological traits, including pathogenic mechanism and resistant mechanism. Moreover, our platform supports studies in pharmaceutical and biotechnology industries, scientific research and agriculture.