Innovation in WGBS: New Methods for DNA Methylation Analysis
Genome-wide methylation sequencing (WGBS) has been a cornerstone in epigenetics research, enabling the exploration of DNA methylation patterns across the genome. However, traditional WGBS has faced limitations, especially in handling low-input samples. In response, novel variants such as PBAT-WGBS, T-WGBS, single-cell WGBS, and nanoporous WGBS have emerged. This review comprehensively examines these technologies, elucidating their principles, advantages, challenges, and potential applications in diverse fields of life science.
Service you may intersted in
PBAT-WGBS
In the vast field of life science, the in-depth exploration of gene expression regulation mechanism has always been the core topic. Epigenetics, as a subject that studies gene expression and functional regulation without DNA sequence changes, has attracted much attention in recent years. WGBS is an important technology in the study of epigenetics DNA methylation, and PBAT-WGBS, as a new technology developed on this basis, is gradually showing its unique advantages and great application potential.
PBAT-WGBS technical principle
- PBAT-WGBS technology is a genome-wide bisulfite sequencing technology based on specific pretreatment and optimization process. Its core principle is still based on WGBS-bisulfite can convert unmethylated cytosine (C) into uracil (U), while methylated cytosine remains unchanged. In the subsequent PCR amplification process, uracil will be amplified into thymine (T). By analyzing the sequencing results and comparing the original sequence, the DNA methylation site can be accurately identified.
- On the basis of traditional WGBS, PBAT-WGBS technology optimizes sample pretreatment and library construction. In the sample pretreatment stage, through specific physical or chemical methods, the genomic DNA is more vulnerable to the action of bisulfite on the premise of maintaining integrity. For example, the genomic DNA is fragmented to a suitable length range by using the gentle ultrasonic fragmentation technology, which not only ensures that bisulfite can fully contact cytosine on the DNA chain, but also avoids the damage to the DNA structure by excessive fragmentation.
- In the aspect of library construction, PBAT-WGBS adopts innovative primer design and linkage strategy. The new primers can bind the DNA fragments treated with bisulfite more efficiently, reduce non-specific amplification and improve the quality and accuracy of library construction. At the same time, the optimized reaction conditions greatly improved the efficiency of connecting the linker with DNA fragments, further ensuring the quality and uniformity of sequencing library.
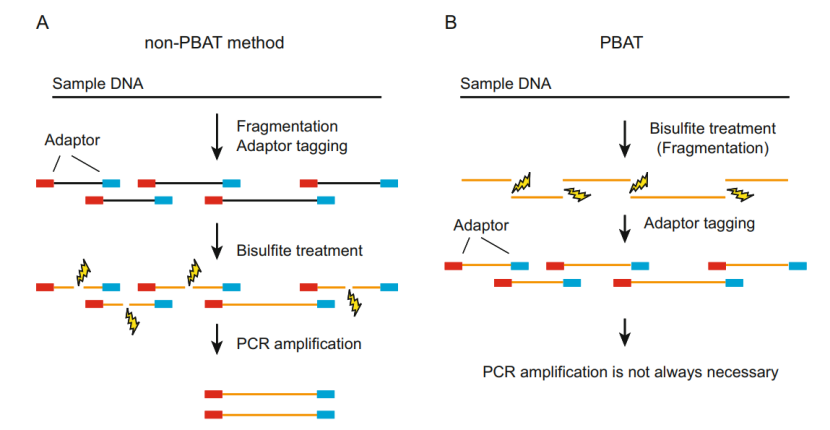 The principle of PBAT-WGBS (Fumihito et al., 2023)
The principle of PBAT-WGBS (Fumihito et al., 2023)
Technical advantages of PBAT-WGBS
- High sensitivity: PBAT-WGBS technology can detect very low levels of DNA methylation changes. In some complex biological processes, such as early embryonic development, small changes in DNA methylation level may have a profound impact on the fate of cells. Traditional methylation detection methods may not capture these subtle changes, but PBAT-WGBS can accurately identify these low-level methylation differences with its optimized pretreatment and sequencing process, which provides a powerful tool for studying the epigenetic regulation mechanism during embryonic development.
- High resolution: This technology can detect DNA methylation with single base resolution. Single base resolution is very important in analyzing methylation patterns of gene promoter regions. Because the methylation status of promoter region is often accurate to a single base, methylation of different base sites may determine whether the gene is expressed. PBAT-WGBS can clearly show the methylation status of each base, which provides the possibility for further study on the fine mechanism of gene expression regulation.
- Wide sample adaptability: PBAT-WGBS technology has relatively loose requirements for samples, and it can be used for effective methylation analysis whether it is precious clinical samples, such as a small amount of tumor puncture tissue or rare species samples that are difficult to obtain. Its optimized sample pretreatment process can retain the methylation information of samples to the maximum extent on the basis of limited sample size, which broadens the application scope of this technology in different fields.
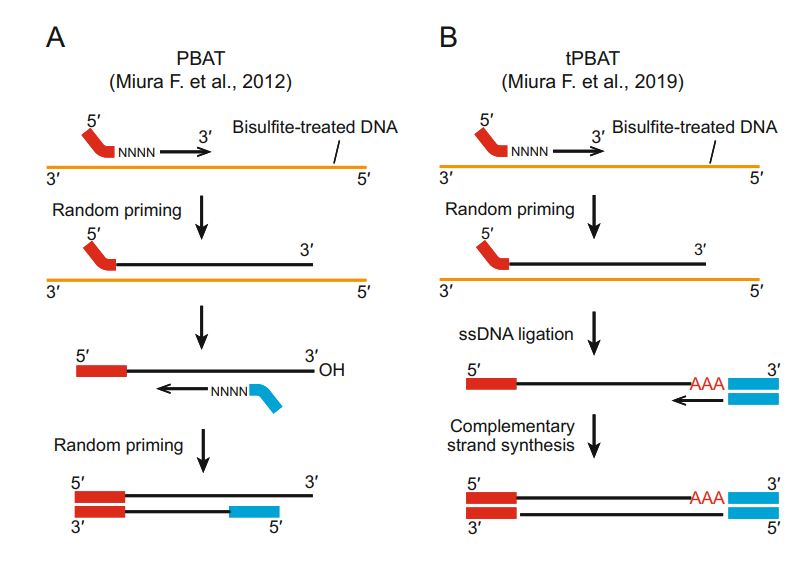 Two PBAT protocols (Fumihito et al., 2023)
Two PBAT protocols (Fumihito et al., 2023)
Challenges and prospects
- Despite its merits, PBAT-WGBS faces challenges. The technology is associated with relatively high costs, encompassing sample processing, reagents, and sequencing expenses. Additionally, the analysis of the massive data generated by PBAT-WGBS requires advanced bioinformatics expertise and high-performance computing resources. Nevertheless, with the continuous evolution of sequencing technology and the development of bioinformatics tools, there is optimism for the future. The decreasing cost of sequencing and the emergence of more efficient data analysis algorithms are expected to enhance the accessibility and utility of PBAT-WGBS in both research and clinical settings. In the long term, its integration with other emerging technologies holds promise for furthering our understanding of life processes and improving disease diagnosis and treatment.
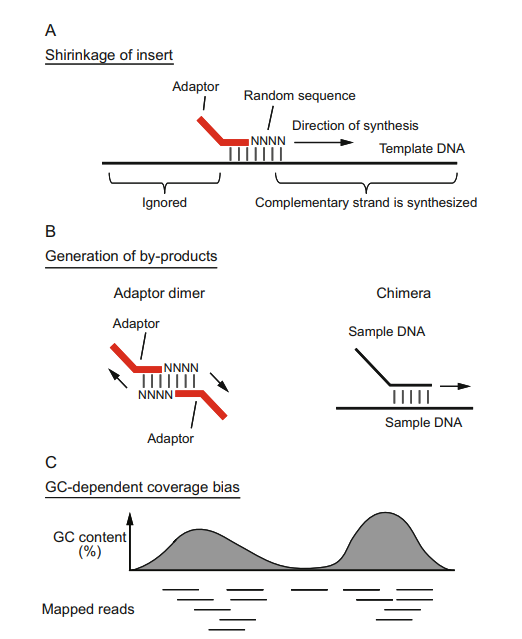 Three drawbacks of random priming PBAT-WGBS (Fumihito et al., 2023)
Three drawbacks of random priming PBAT-WGBS (Fumihito et al., 2023)
T-WGBS
In life science research, DNA methylation, as an important epigenetic modification, plays a key role in gene expression regulation, cell differentiation and disease occurrence and development. WGBS is the gold standard method to detect DNA methylation, and targeted whole genome bisulfite sequencing (T-WGBS) is an innovative technology developed on this basis.
- WGBS technology aims at high-depth methylation analysis of specific regions of genome. It combines the basic principle of bisulfite transformation, that is, bisulfite can transform unmethylated cytosine into uracil, while methylated cytosine remains unchanged. After subsequent PCR amplification and sequencing, the methylation site is determined by comparison with the reference genome. Different from the traditional WGBS, T-WGBS focuses its research on the genome region of interest through targeted enrichment strategy, thus reducing the sequencing cost and significantly improving the sequencing depth and data accuracy of the target region.
T-WGBS technology principle
- Targeted region selection: Firstly, according to the research purpose, researchers select specific genomic regions, such as gene promoters, enhancers, CpG islands and other regions closely related to gene expression regulation with the help of bioinformatics tools. The methylation status of these regions often has an important influence on gene function.
- DNA fragmentation: The genomic DNA is cut into fragments with appropriate size by physical methods (such as ultrasonic fragmentation) or enzyme digestion, with a general length of about 100-500bp for subsequent operation.
- Targeted enrichment: Probe hybridization or PCR-based method is used to enrich DNA fragments in the target area. Probe hybridization is to design a DNA or RNA probe complementary to the target region, and capture the target fragment through liquid phase or solid phase hybridization. The enrichment based on PCR is to design specific primers for the target region and amplify the target fragment.
- Bisulfite transformation: The enriched DNA fragments are treated with bisulfite to transform unmethylated cytosine into uracil, while methylated cytosine is not affected. This step is the key to realize methylation detection, and the transformation efficiency directly affects the accuracy of subsequent results.
- Library construction and sequencing: The transformed DNA fragments were subjected to terminal repair, A-tail addition, sequencing linker connection and other operations to construct a sequencing library. Subsequently, the second-generation sequencing technology (such as Illumina platform) was used to high-throughput sequencing the library to obtain a large number of sequence data containing methylation information.
- Data analysis: Through bioinformatics algorithm, the sequencing data were compared with the reference genome, the methylation sites were identified, and the methylation level was quantitatively analyzed. At the same time, combined with relevant biological databases, the biological significance of methylation and gene expression, disease association and so on was explored.
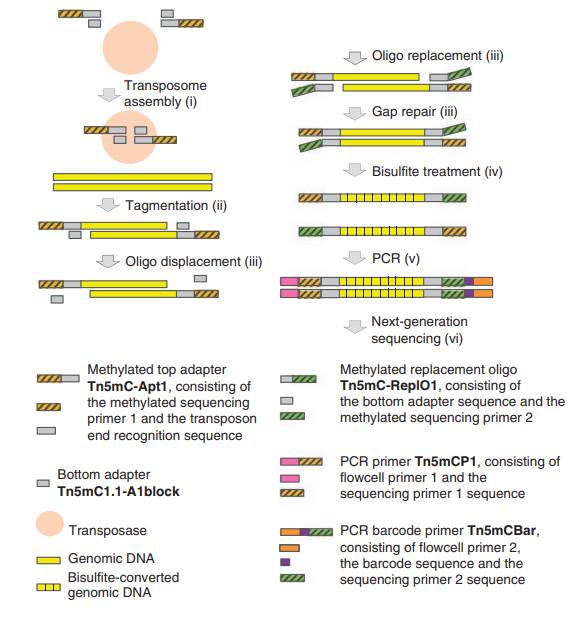 Overview and components of T-WGBS library preparation (Wang et al., 2022)
Overview and components of T-WGBS library preparation (Wang et al., 2022)
Technical advantages of T-WGBS
- Cost-effective: Compared with traditional WGBS, T-WGBS only targets specific regions, which greatly reduces the amount of sequencing data and the sequencing cost, and enables researchers to conduct more in-depth methylation research within a limited budget.
- High-depth sequencing: Because the sequencing data are concentrated in the target areas, T-WGBS can achieve high-depth coverage of these areas, usually reaching hundreds or even thousands of times the sequencing depth. This makes it possible to detect low-level methylation signals and methylation differences in rare cell populations, and improves the sensitivity and accuracy of detection.
- Strong flexibility: researchers can flexibly select different targeted regions according to their own research needs, whether it is a specific gene family, a specific chromosome segment, or a candidate gene region related to diseases, and can conduct targeted analysis to meet diversified research needs.
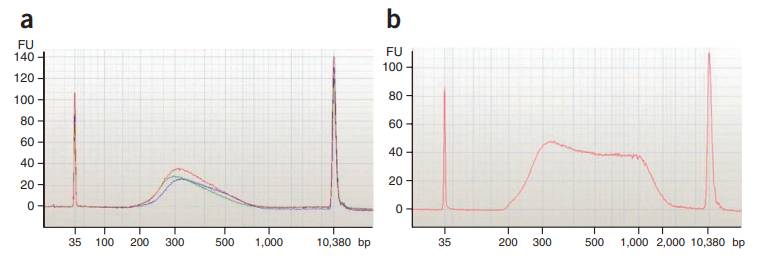 Size distribution of T-WGBS libraries (Wang et al., 2022)
Size distribution of T-WGBS libraries (Wang et al., 2022)
Challenges and prospects
- T-WGBS is not without challenges. In the targeted enrichment process, issues such as non-specific enrichment and variable enrichment efficiency may arise, potentially compromising the accuracy and reliability of the data. Additionally, although the data is focused on specific regions, the high-depth sequencing still generates a substantial amount of data that demands sophisticated bioinformatics knowledge and tools for proper interpretation. Looking ahead, efforts are underway to optimize the targeted enrichment technology by developing improved probe design strategies and enrichment methods. Concurrently, the bioinformatics field is actively engaged in creating more efficient data analysis algorithms and tools to facilitate the utilization of T-WGBS data by a broader range of researchers. With these advancements, T-WGBS is expected to play an increasingly important role in elucidating the role of DNA methylation in biological processes and advancing precision medicine and agricultural biotechnology.
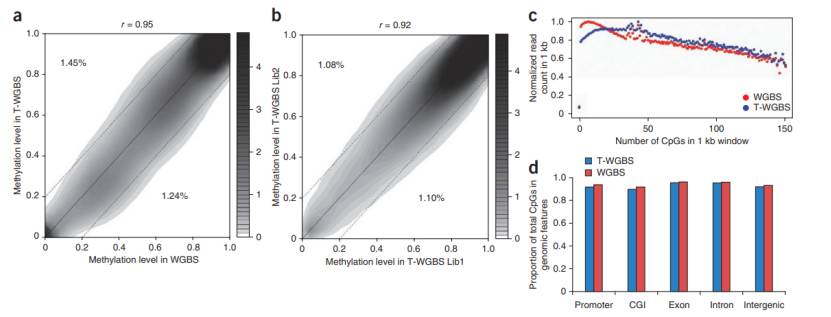 Reliability and reproducibility of T-WGBS (Wang et al., 2022)
Reliability and reproducibility of T-WGBS (Wang et al., 2022)
Single-cell WGBS
In life science research, cells are not homogeneous groups, and each cell has unique molecular characteristics. The traditional WGBS technology analyzes a large number of cells, and the result is the average methylation level of the cell population, which masks the methylation differences between cells. However, the heterogeneity of DNA methylation between cells is very important in the process of embryo development and tumor occurrence and development. For example, in the early stage of embryonic development, the determination of cell fate is closely related to the dynamic changes of DNA methylation, and there are significant differences between cells. In order to deeply explore the epigenetic information of individual cells, single cell WGBS technology came into being.
Single-cell WGBS technical principle
- Single cell separation: Accurately obtaining a single cell from tissue or cell suspension is the first step of single cell WGBS. Commonly used methods are fluorescence activated cell sorting (FACS), which sorts specific cells according to the fluorescence signals of cell surface markers. There is also a micromanipulation method, which directly selects target single cells with the help of a microscope. Microfluidic technology realizes efficient capture and separation of single cells through microchannels and microvalves.
- DNA amplification: Because the DNA content of a single cell is very small, it is necessary to amplify the whole genome. Multiplex displacement amplification (MDA) is a common method. With random primers and Phi29 DNA polymerase with strand displacement activity, the whole genome can be amplified at constant temperature, and microgram DNA products can be obtained to meet the needs of subsequent experiments. However, MDA also has the problem of amplification deviation, which leads to excessive or insufficient amplification in some areas.
- Transformation of bisulfite: Like traditional WGBS, unmethylated cytosine is converted into uracil by bisulfite, while methylated cytosine remains unchanged. This step requires high transformation efficiency, because the amount of DNA in a single cell is small, and if the transformation is incomplete, it will seriously affect the accurate identification of methylation sites.
- Library construction and sequencing: The transformed DNA was subjected to terminal repair, A-tail addition, sequencing linker connection and other operations to construct a library, and then high-throughput sequencing was performed by the second generation sequencing technology (such as Illumina platform) to obtain sea sequencing data.
- Data analysis: By using bioinformatics algorithm, the sequencing data were compared with the reference genome, the methylation sites were identified, the methylation patterns of single cells were analyzed, and the differences of methylation between cells and their biological significance were explored.
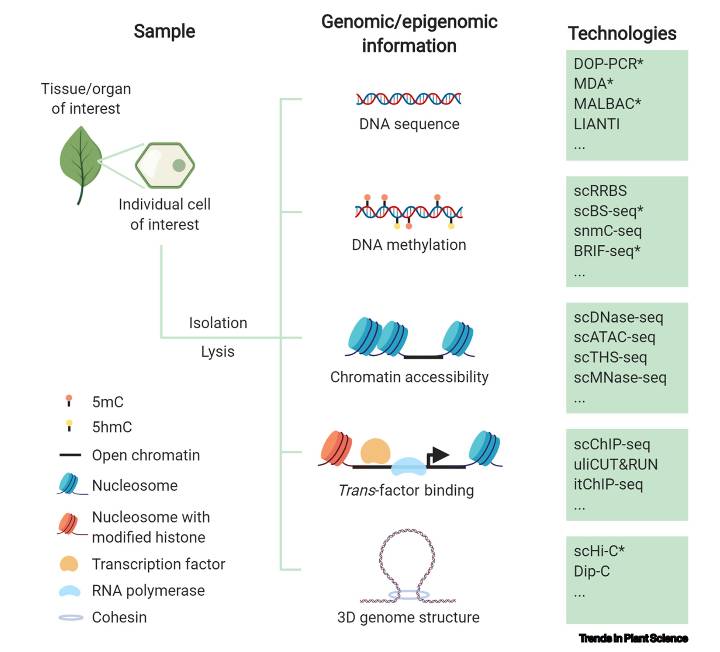 Genomic and epigenomic information can be interrogated in a single cell via multiple single-cell sequencing technologies (Luo et al., 2020)
Genomic and epigenomic information can be interrogated in a single cell via multiple single-cell sequencing technologies (Luo et al., 2020)
Technical advantages of single-cell WGBS
- Reveal the heterogeneity of cells: It can show the DNA methylation state of a single cell and reveal the hidden methylation heterogeneity in the cell population. In tumor research, methylation differences among tumor cell subsets can be found, which is helpful to understand the heterogeneity and evolution of tumors.
- The study of rare cells: Methylation analysis of rare cells such as a few cells and circulating tumor cells in early embryonic development provides key information for related research.
- Dynamic monitoring of cell changes: In the process of cell differentiation, disease development, etc., we can dynamically track the changes of methylation in individual cells and deeply understand the epigenetic mechanism of cell state transition.
Challenges and prospects
- Single-cell WGBS faces several technical and analytical challenges. Technically, the low amount of starting DNA in a single cell makes it vulnerable to contamination, and amplification biases can affect data accuracy. Additionally, ensuring the efficient construction of libraries from low-concentration DNA is a nontrivial task. From an analytical perspective, the large amount of data generated, combined with high noise levels, necessitates the development of advanced algorithms and tools for meaningful data extraction. Nevertheless, ongoing technological innovations are expected to overcome these hurdles. The development of more precise single cell isolation techniques, improved DNA amplification methods with reduced biases, and enhanced bisulfite transformation protocols will enhance the reliability of single-cell WGBS. Concurrently, the evolution of powerful data analysis methods will enable more comprehensive interpretation of the data, thereby driving significant advancements in life science research.
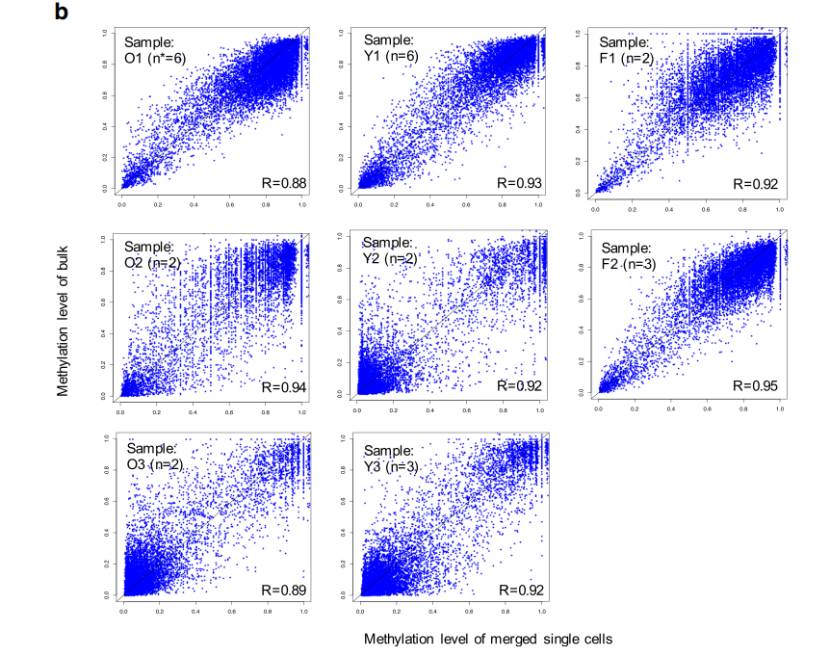 Single-cell WGBS is an accurate and reproducible method for genome-wide 5mC analysis (Luo et al., 2020)
Single-cell WGBS is an accurate and reproducible method for genome-wide 5mC analysis (Luo et al., 2020)
Service you may intersted in
Nano-porous WGBS
In the field of life science, DNA methylation, as a key epigenetic modification, has a far-reaching impact on many processes such as gene expression regulation, cell differentiation and disease occurrence and development. WGBS has always been an important means to detect DNA methylation, and nano-porous WGBS, as a new technology, is gradually emerging, bringing new opportunities for research in this field.
Nano-porous WGBS technical principle
- Nano-pore sequencing is based on nano-scale pores. When DNA molecules pass through the nano-pores under the action of external electric field, it will cause the change of ion current in the pores. Because different bases or modified bases have different effects on ion current, the determination of DNA sequence and modification can be realized by monitoring the current change in real time.
- In nano-porous WGBS, bisulfite treatment is still the key step. After bisulfite treatment, unmethylated cytosine is converted into uracil, while methylated cytosine remains unchanged. Subsequently, the treated DNA passes through the nanopore, and the methylated cytosine and unmethylated uracil have different characteristics of ion current when passing through the nanopore, thus distinguishing methylation sites. Compared with traditional sequencing technology, nanopore sequencing can directly read DNA sequence and methylation information without PCR amplification and fluorescence labeling, which reduces the deviation introduced by amplification.
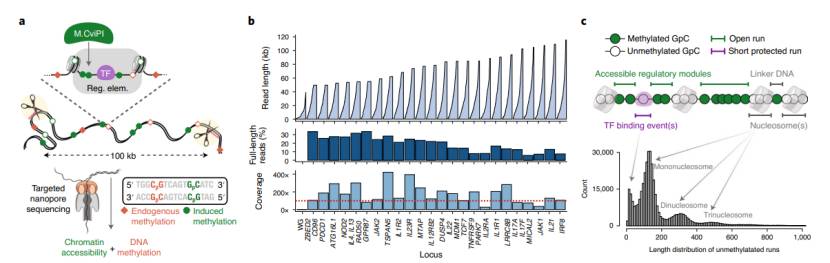 Phasing chromatin accessibility and DNA methylation across long single molecules (Sofia et al., 2022)
Phasing chromatin accessibility and DNA methylation across long single molecules (Sofia et al., 2022)
Technical advantages of nano-porous WGBS
- Real-time detection: Nanoporous WGBS can monitor the process of DNA passing through the nanopore in real time and obtain methylation information in real time. In tumor research, the dynamic monitoring of methylation status of tumor patients during treatment can reflect the treatment effect in time and provide a basis for adjusting the treatment plan.
- Long reading length: Nanopore sequencing can produce long reading length sequences, which is very important for analyzing methylation in complex genomic regions. In some regions with highly repetitive sequences, long reading length can better determine methylation patterns, while short reading length sequencing technology may lose important information in the splicing process. Direct detection of methylation: The methylation status can be inferred without complicated subsequent modification or amplification of DNA treated with bisulfite, and methylated cytosine can be directly identified from the change of ion current, thus avoiding the errors introduced by additional experimental steps and improving the accuracy of methylation detection.
- Portable and on-site detection: Nanopore sequencing equipment is relatively small and portable, and can be used for on-site sample detection. In clinical diagnosis, it can quickly provide test results for patients, especially for remote areas or emergency medical scenes.
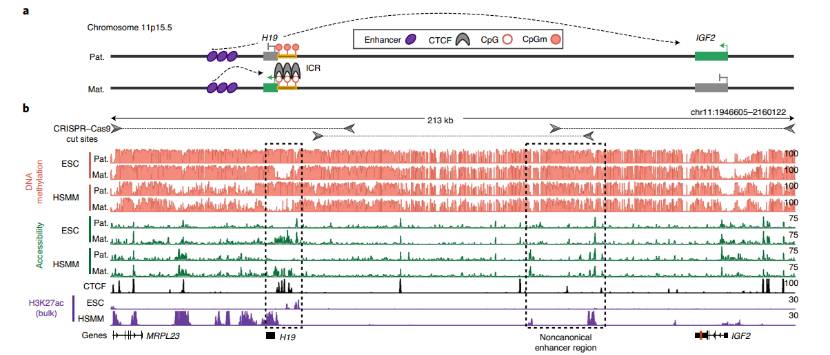 Phased epigenomic profiles deconvolve alternate H19/IGF2 alleles (Sofia et al., 2022)
Phased epigenomic profiles deconvolve alternate H19/IGF2 alleles (Sofia et al., 2022)
Challenges and prospects
- Despite its potential, nano-porous WGBS faces challenges. In terms of signal interpretation, the complex ion current signals require advanced analytical methods, and the application of artificial intelligence and machine learning algorithms is being explored to improve the accuracy of methylation site identification. To enhance the technology's practicality, efforts are focused on increasing sequencing throughput and reducing costs through the development of new nanoporous materials and manufacturing processes. Additionally, optimizing the sample treatment process, particularly the bisulfite treatment method, is crucial for improving the overall performance of the technology. With continued research and development, nano-porous WGBS is expected to make significant contributions to epigenetics research and have a profound impact on the fields of life science and medicine.
The development of novel WGBS technologies, including PBAT-WGBS, T-WGBS, single-cell WGBS, and nano-porous WGBS, represents a significant step forward in the field of epigenetics. These techniques address the limitations of traditional WGBS and offer unique advantages in terms of sensitivity, resolution, cost-effectiveness, and sample adaptability. While each technology faces its own set of challenges, ongoing research efforts and technological advancements are expected to overcome these obstacles.
References
-
Luo Cheng, Alisdair R. Fernie and Yan Jianbing. "Single-Cell Genomics and Epigenomics: Technologies and Applications in Plants." Trends in Plant Science (2020) 1031-1040. https://doi.org/10.1016/j.tplants.2020.04.016
- Slivia Gravina, Dong Xiao, Yu Bo and Jan Vijg. Single-cell genome-wide bisulfite sequencing uncovers extensive heterogeneity in the mouse liver methylome." Genome Biology (2016) 17:150. DOI 10.1186/s13059-016-1011-3
- Wang Qi, Gu Lei, Andrew Adey, Wang Wei and Dieter Weichenhan. "Tagmentation-based whole-genome bisulfite sequencing." Nature Protocols (2013): 2022-2032. doi:10.1038/nprot.2013.118
- Fumihito Miura, Yukiko Shibata, Miki Miura and Takashi Ito. "Post-bisulfite Adaptor Tagging Based on an ssDNA Ligatio Technique (tPBAT)." Epigenomics (2023): 2577. https://doi.org/10.1007/978-1-0716-2724-2_2
- Sofia Battaglia, Kevin Dong, Wu Jingyi, Chen Zeyu and Bradley E. Bernstein. "Long-range phasing of dynamic, tissue-specific and allele-specific regulatory elements." Nature Genetics (2022): 1504–1513. https://doi.org/10.1038/s41588-022-01188-8
! For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.

 The principle of PBAT-WGBS (Fumihito et al., 2023)
The principle of PBAT-WGBS (Fumihito et al., 2023) Two PBAT protocols (Fumihito et al., 2023)
Two PBAT protocols (Fumihito et al., 2023) Three drawbacks of random priming PBAT-WGBS (Fumihito et al., 2023)
Three drawbacks of random priming PBAT-WGBS (Fumihito et al., 2023) Overview and components of T-WGBS library preparation (Wang et al., 2022)
Overview and components of T-WGBS library preparation (Wang et al., 2022) Size distribution of T-WGBS libraries (Wang et al., 2022)
Size distribution of T-WGBS libraries (Wang et al., 2022) Reliability and reproducibility of T-WGBS (Wang et al., 2022)
Reliability and reproducibility of T-WGBS (Wang et al., 2022) Genomic and epigenomic information can be interrogated in a single cell via multiple single-cell sequencing technologies (Luo et al., 2020)
Genomic and epigenomic information can be interrogated in a single cell via multiple single-cell sequencing technologies (Luo et al., 2020) Single-cell WGBS is an accurate and reproducible method for genome-wide 5mC analysis (Luo et al., 2020)
Single-cell WGBS is an accurate and reproducible method for genome-wide 5mC analysis (Luo et al., 2020) Phasing chromatin accessibility and DNA methylation across long single molecules (Sofia et al., 2022)
Phasing chromatin accessibility and DNA methylation across long single molecules (Sofia et al., 2022) Phased epigenomic profiles deconvolve alternate H19/IGF2 alleles (Sofia et al., 2022)
Phased epigenomic profiles deconvolve alternate H19/IGF2 alleles (Sofia et al., 2022)