The Role of DNA Methylation in Modern Biomedical Research
DNA methylation, as one of the core mechanisms of epigenetics, plays a crucial role in modern biomedical research. This biochemical process adds methyl groups to DNA molecules, regulating gene expression without altering the DNA sequence itself, providing the organism with flexible and precise gene regulation mechanisms. With the rapid development of high-throughput sequencing technology, research into DNA methylation has deepened, revealing its widespread involvement and critical role in genetic imprinting, embryonic development, cell differentiation, and the development and progression of diseases.
In the field of biomedicine, DNA methylation is not only an important window for understanding gene function, cell fate determination, and disease mechanisms, but also provides new perspectives and strategies for early diagnosis, prognosis assessment, and personalized treatment of diseases. For example, changes in the methylation pattern of specific genes have become molecular markers for various cancers, allowing doctors to detect tumor signs earlier and develop more precise treatment plans. Moreover, DNA methylation is closely related to the occurrence and development of complex diseases such as neurodegenerative diseases and cardiovascular diseases, and its research is expected to open up new avenues for the prevention and treatment of these diseases.
In summary, DNA methylation, a hot topic and cutting-edge research in modern biomedical studies, not only helps unveil the mysteries of life sciences but also brings hope and challenges for the prevention and treatment of human health and diseases.
DNA Methylation in Modern Biological Research
DNA methylation refers to the process in which the 5' end of the CpG island in the genome DNA sequence is converted from cytosine to 5'-methylcytosine (5mC) under the action of DNA methyltransferase (DNMT). This modification method does not change the gene sequence, but it can affect the expression pattern of genes. DNA methylation usually occurs in CpG dinucleotides and is one of the main epigenetic forms of gene expression regulation in mammals. As an important epigenetic modification, DNA methylation plays a crucial role in modern biomedical research.
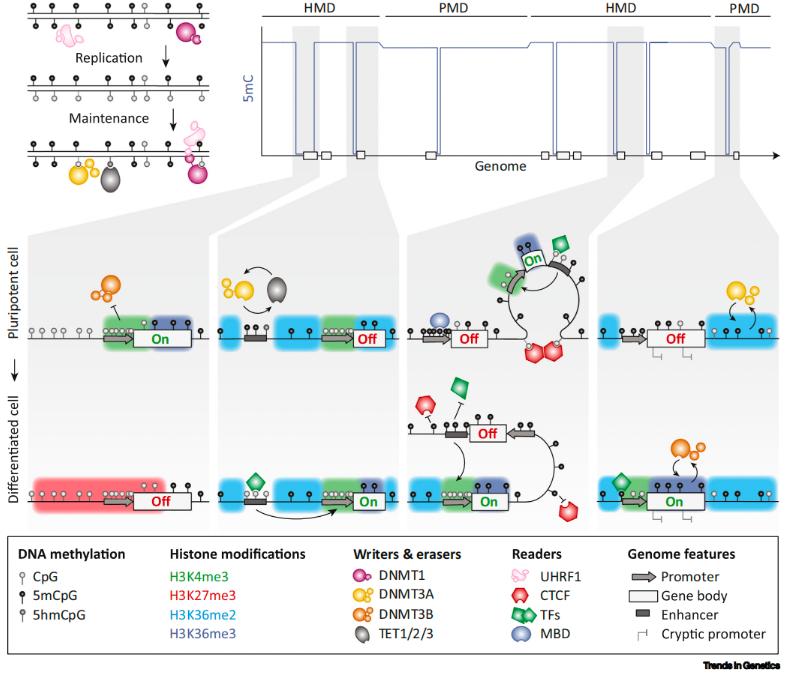 Key concepts of DNA methylation gene regulation (Mattei et al., 2022)
Key concepts of DNA methylation gene regulation (Mattei et al., 2022)
What are the types of DNA methylation?
- De novo methylation: De novo methylation refers to the process of adding methyl groups to entirely new DNA sites without relying on existing methylated DNA strands. This type of methylation primarily occurs during the early stages of embryonic development and in certain cell reprogramming processes. It involves specific methyltransferases, such as Dnmt3a and Dnmt3b, which can add methyl groups to DNA molecules in the absence of a pre-existing methylation template. Clinically, this mechanism is applicable in studies related to embryonic development, cellular differentiation, and cancer research.
- Maintenance methylation: Maintenance methylation describes the process by which one strand of double-stranded DNA that has already been methylated leads to the corresponding unmethylated strand being modified during DNA replication, thereby preserving the original pattern of methylation. This form of methylation is primarily catalyzed by maintenance methyltransferase (such as Dnmt1), which accurately replicates the established patterns from parental DNA strands onto daughter strands during replication, ensuring stable transmission of genetic information. Clinically, it holds significance for researching hereditary diseases.
Biological Research on DNA Methylation
- Regulation of Gene Expression: DNA methylation regulates gene expression by altering chromatin structure, DNA conformation, and the way DNA interacts with proteins. In eukaryotic organisms, approximately 60%-90% of CpG dinucleotides in the genome display methylation of cytosine, which can inhibit or activate the expression of specific genes.
- Genome Stability and Genetic Imprinting: DNA methylation is crucial for maintaining genome stability. Research has shown that abnormal DNA methylation states are closely related to the development and progression of various diseases, especially cancer. In addition, DNA methylation also plays a role in the transmission of genetic imprinting, which has important implications for embryonic development and the formation of genetic traits.
- Immune Response and Defense Mechanisms: DNA methylation also plays an important role in the immune response. Bacterial DNA is mostly unmethylated, while eukaryotic DNA is mostly methylated. When the body detects unmethylated DNA, it will regard it as an invading foreign substance and trigger an immune response. This mechanism helps the body resist the invasion of foreign pathogens.
What is the effect of DNA methylation on gene expression?
Directly affecting gene transcription
- Preventing transcription factor binding: DNA methylation status can directly prevent transcription factors from binding to the promoter regions of genes. In methylated DNA sites, methyl groups attract methylation binding proteins (such as MBD proteins), which form tight chromatin complexes with methylated DNA, further preventing the approach of transcription factors and thus inhibiting gene expression.
- Changing chromatin structure: DNA methylation can also affect gene expression by changing chromatin structure indirectly. Methylated DNA sites promote the formation of tighter chromatin structures, such as heterochromatin, which is unfavorable for the binding of transcription factors and RNA polymerase, thus inhibiting gene expression.
Indirectly affecting gene expression
- Regulating transcription factor activity: DNA methyltransferases (such as DNMTs) can interact with transcription factors and regulate their activity and degradation rate. This interaction may affect the stability of transcription factors or their ability to bind to other regulatory factors, indirectly affecting the expression level of genes.
- Influencing chromatin three-dimensional structure: Methylated DNA sites may also affect gene expression by further influencing chromatin's three-dimensional structure and interactions between chromatin. This influence may involve advanced folding of chromatin, nucleosome positioning, and the formation of chromatin domains at multiple levels.
Service you may intersted in
What are the medical applications of DNA methylation?
DNA methylation, as an important epigenetic modification, has broad application prospects in modern biomedical research. It not only regulates gene expression, maintains genome stability, but also participates in multiple biological processes, including disease development and immune responses. With the continuous advancement of research techniques and further exploration, DNA methylation is sure to play a more important role in disease diagnosis, drug development, and regenerative medicine.
Disease diagnosis and risk assessment
- Early diagnosis: DNA methylation patterns change significantly during disease states, and these changes often precede the appearance of clinical symptoms. Therefore, by detecting the methylation level of specific genes, early diagnosis of diseases can be achieved. For example, in the early detection of cancer, the hypermethylated state of certain tumor suppressor genes can serve as a biomarker for cancer, helping doctors spot signs of cancer before patients show obvious symptoms. The following picture depicts a prospective population-based cohort study of more than 1200 children with newly diagnosed CNS tumors. Combining methylomics, targeted genome sequencing, and a reference diagnostic approach to neuropathology, to evaluate the role of DNA methylation analysis in the diagnosis of CNS tumors in children and adolescents. This study provides a basis for the application of DNA methylation in the neuropathological diagnosis of central nervous system tumors in children and adolescents.
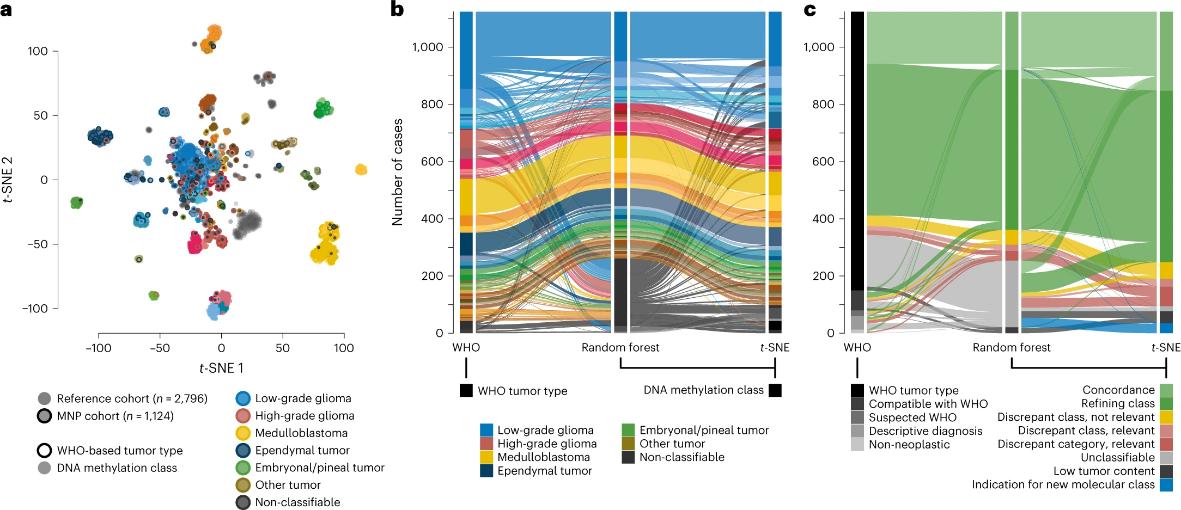 Application of DNA methylation in neuropathological diagnosis (Sturm et al., 2023)
Application of DNA methylation in neuropathological diagnosis (Sturm et al., 2023)
- Risk assessment: Changes in DNA methylation levels can also be used to assess an individual's risk of developing a disease. For example, by testing the level of methylation of specific genes in the blood, a person's risk of developing Alzheimer's disease can be assessed. Studies have shown that methylation levels of certain genes are associated with the onset and progression of Alzheimer's disease, and abnormal methylation levels of these markers in the blood may be associated with an increased risk of Alzheimer's disease.
Evaluation of disease prognosis and curative effect
- Prognostic assessment: Changes in DNA methylation levels can also reflect the prognosis of the disease. For example, in cancer patients, the methylation pattern of specific genes may be associated with prognostic indicators such as patient survival and risk of recurrence. By measuring the methylation levels of these genes, doctors can assess a patient's prognosis and provide a basis for personalized treatment.
- Evaluation of efficacy: During treatment, changes in DNA methylation can reflect the therapeutic effect. For example, certain tumors may show changes in DNA methylation patterns after chemotherapy, and these changes can be used to assess the effects of chemotherapy. If the treatment causes the DNA methylation pattern to shift to a normal state, this may indicate the effectiveness of the treatment; conversely, if the methylation pattern does not change or if further abnormalities appear, the treatment regimen may need to be adjusted.
Drug development and personalized treatment
- Drug target screening: Genes with abnormal DNA methylation or their regulatory networks can be used as potential targets for therapy. For example, targeting highly methylated tumor suppressor genes in certain cancers, demethylation drugs can be used to reactivate the expression of these genes, thereby inhibiting tumor growth. Through big data analysis and bioinformatics tools, researchers can identify methylation markers associated with specific diseases, providing targets for precision medicine.
- Drug development: Based on the regulatory mechanism of DNA methylation, novel therapeutic drugs can be developed. These drugs achieve therapeutic goals by inhibiting the activity of DNA methyltransferase (DNMT), reducing abnormal DNA methylation levels and restoring the expression of tumor suppressor genes. With the development of science and technology, especially in the fields of epigenetics, genomics and bioinformatics, the development of drugs based on DNA methylation will be more accurate and efficient. Researchers focused on DNA methylation and found the novel DNMT1/HDAC dual inhibitor (R)-23a effectively reverses the cancer-specific epigenetic abnormalities and holds great potential for further development into cancer therapeutic agents.
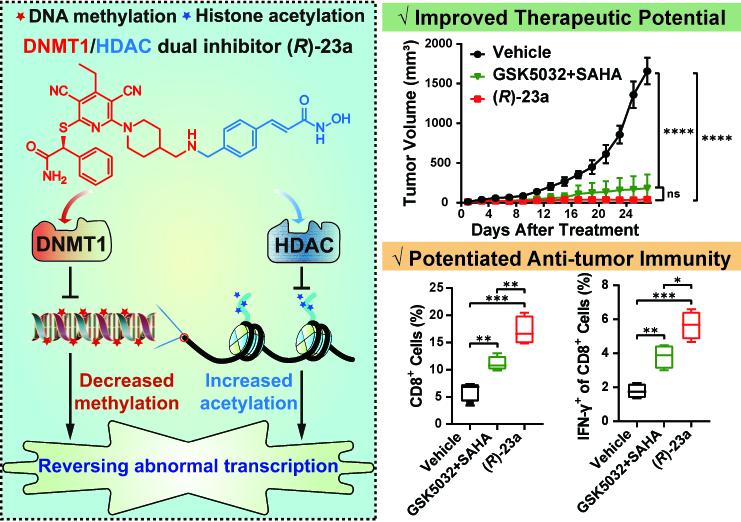 DNMT1/HDAC dual-acting inhibitor (R)-23a design strategy and activity data (Chang et al., 2024)
DNMT1/HDAC dual-acting inhibitor (R)-23a design strategy and activity data (Chang et al., 2024)
Research on embryonic development and regenerative medicine
- Regulation of embryonic development: DNA methylation plays an important role in embryonic development, it is involved in the regulation of cell differentiation and tissue-specific gene expression. By studying the changes of DNA methylation in different stages of embryonic development, we can reveal the molecular mechanism of embryonic development and provide theoretical support for the development of regenerative medicine.
- Regulation of stem cell differentiation: DNA methylation also plays a key role in stem cell research. By regulating the methylation state of stem cells, stem cells can be guided to differentiate in a specific direction, providing new ideas and methods for tissue engineering and regenerative medicine.
Bioinformatics for DNA Methylation
Methylation Patterns and Levels
Methylation Site Identification: This analysis identifies which cytosine (C) residues in the genome have methyl (-CH3) groups added, with a particular focus on CpG dinucleotides.
Methylation Level: The methylation ratio for each CpG site is calculated as the proportion of methylated reads to total reads, providing insights into the methylation levels at various sites.
 Methylation Site Identification of Cancer (Liang et al., 2023)
Methylation Site Identification of Cancer (Liang et al., 2023)
Differential Methylation Analysis
Differential Methylation Sites (DMCs): Statistical methods such as t-tests, Wilcoxon rank-sum tests, and linear models are employed to compare differences in methylation levels across different samples or groups, thereby identifying differential methylation sites.
Differential Methylation Regions (DMRs): Further examination of these differential methylation sites allows for the identification of regions that may be associated with specific biological processes or disease states.
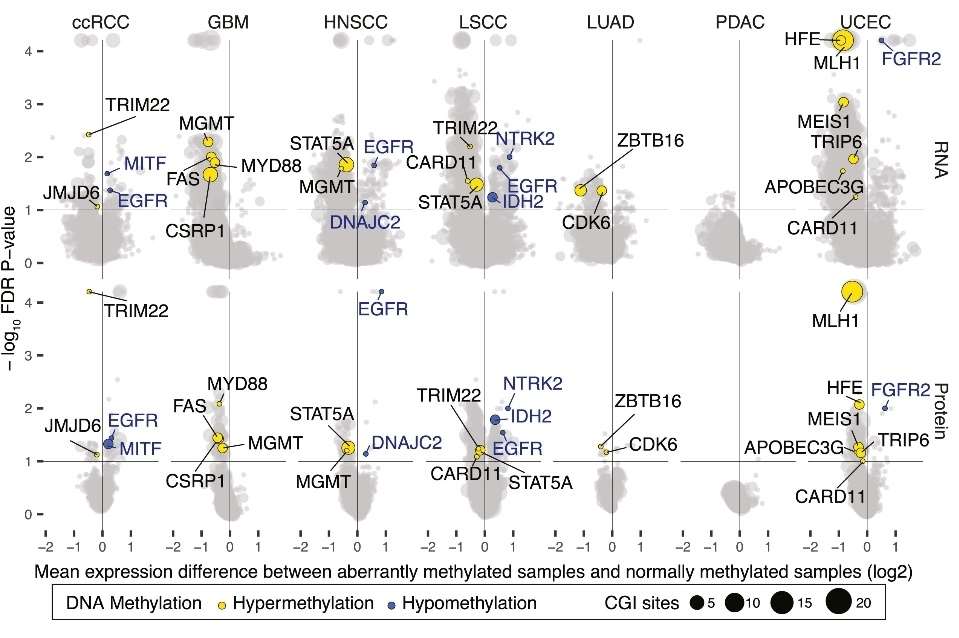
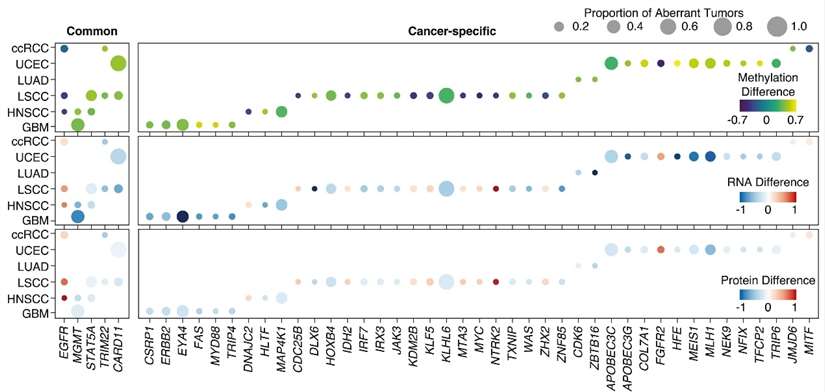 The median methylation, RNA, and protein differences for cancer-associated genes (Liang et al., 2023)
The median methylation, RNA, and protein differences for cancer-associated genes (Liang et al., 2023)
Functional Annotation and Biological Significance
Gene Ontology (GO) Analysis: Mapping differential methylation sites onto the genome facilitates an understanding of their corresponding genes and functions, revealing their roles in biological processes through GO analysis.
Gene Set Enrichment Analysis (GSEA): This analysis assesses whether differential methylation sites are enriched within specific gene sets or pathways to uncover underlying molecular mechanisms.
Pathway Analysis
Gene Pathway Analysis: Tools such as KEGG and Reactome are utilized to analyze gene pathways associated with differential methylation sites, elucidating interactions and regulatory relationships among these genes within biological contexts.
The Relationship Between Methylation and Gene Expression: By integrating gene expression data, this analysis explores how changes in DNA methylation influence gene expression dynamics.
Visual Display
Heatmap: Displays variations in methylation levels between different samples or groups along with the distribution of differential methylated sites.
Volcano Plot: Illustrates both statistical significance and effect size for differential methylted sites, aiding researchers in swiftly identifying critical locations.
Manhattan Plot: Depicts the genomic distribution of differential methylted sites to facilitate identification of variants linked to specific biological processes or diseases.
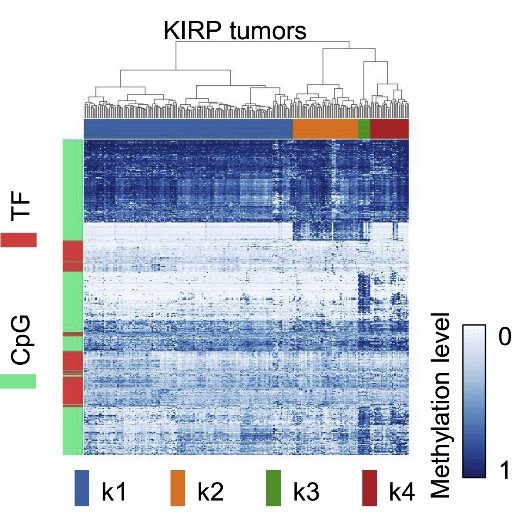 Heatmap of CpG site methylation and TF expression profiles (Liu et al., 2019)
Heatmap of CpG site methylation and TF expression profiles (Liu et al., 2019)
References
- Chen, Yang, En-Min Li, and Li-Yan Xu. "Guide to metabolomics analysis: a bioinformatics workflow." Metabolites 12.4 (2022): 357.
- Mattei AL, Bailly N, Meissner A. DNA methylation: a historical perspective. Trends Genet. 2022;38(7):676-707.
- Sturm, D., Capper, D., Andreiuolo, F. et al. Multiomic neuropathology improves diagnostic accuracy in pediatric neuro-oncology. Nat Med 29, 917–926 (2023).
- Chang Y, Guo H, Li X, et al. Development of a First-in-Class DNMT1/HDAC Inhibitor with Improved Therapeutic Potential and Potentiated Antitumor Immunity. J Med Chem. 2024;67(18):16480-16504.
- Liang WW, Lu RJ, Jayasinghe RG, et al. Integrative multi-omic cancer profiling reveals DNA methylation patterns associated with therapeutic vulnerability and cell-of-origin. Cancer Cell. 2023;41(9):1567-1585.e7.
- Liu Y, Liu Y, Huang R, et al. Dependency of the Cancer-Specific Transcriptional Regulation Circuitry on the Promoter DNA Methylome. Cell Rep. 2019;26(12):3461-3474.e5.
! For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.

 Key concepts of DNA methylation gene regulation (Mattei et al., 2022)
Key concepts of DNA methylation gene regulation (Mattei et al., 2022)  Application of DNA methylation in neuropathological diagnosis (Sturm et al., 2023)
Application of DNA methylation in neuropathological diagnosis (Sturm et al., 2023)  DNMT1/HDAC dual-acting inhibitor (R)-23a design strategy and activity data (Chang et al., 2024)
DNMT1/HDAC dual-acting inhibitor (R)-23a design strategy and activity data (Chang et al., 2024)  Methylation Site Identification of Cancer (Liang et al., 2023)
Methylation Site Identification of Cancer (Liang et al., 2023)  The median methylation, RNA, and protein differences for cancer-associated genes (Liang et al., 2023)
The median methylation, RNA, and protein differences for cancer-associated genes (Liang et al., 2023)  Heatmap of CpG site methylation and TF expression profiles (Liu et al., 2019)
Heatmap of CpG site methylation and TF expression profiles (Liu et al., 2019)