We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) is a powerful technique in epigenomics, allowing researchers to study chromatin accessibility. ATAC-seq has broad applications in biomedical research, ranging from disease pathogenesis to drug discovery, and it plays a key role in understanding cellular differentiation at single-cell resolution. Below is a detailed exploration of the applications of ATAC-seq in various fields.
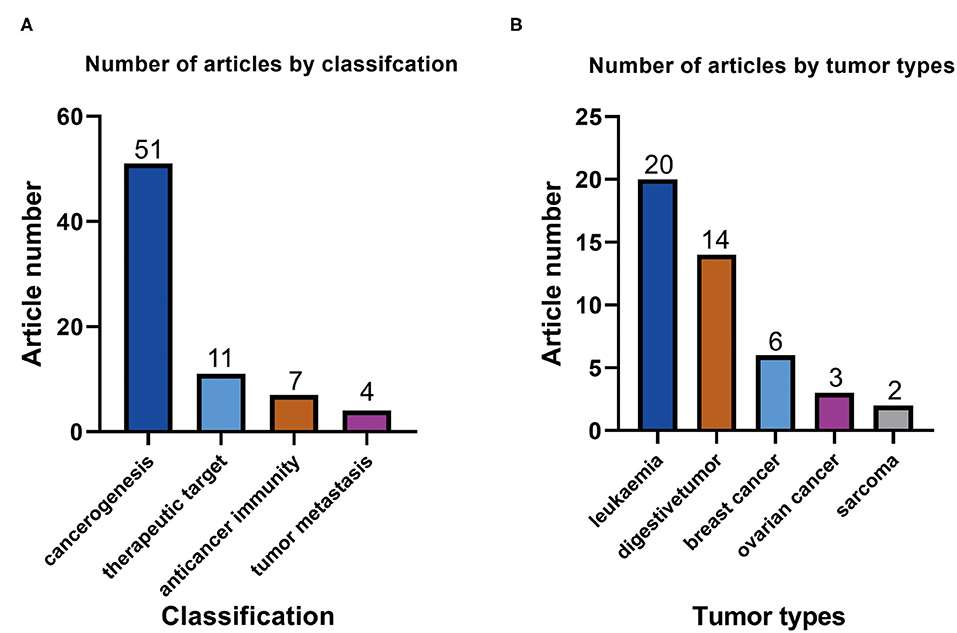 Number of ATAC-seq-related articles with different classifications1 (Zhao, Y. et al,2020)
Number of ATAC-seq-related articles with different classifications1 (Zhao, Y. et al,2020)
Chromatin accessibility is crucial for gene regulation, and changes in chromatin structure are often linked to diseases. ATAC-seq helps reveal which regions of the genome are open and available for transcription, providing valuable insights into how epigenetic mechanisms contribute to health and disease2.
In complex diseases such as cardiovascular disorders or diabetes, ATAC-seq can identify open chromatin regions associated with specific gene regulation changes, offering clues to genetic susceptibility. For example, Davidson, R. K. et al used ATAC-seq to identify chromatin regions associated with insulin regulation, linking them to gene expression changes that impact diabetes-related β-cell function3. Additionally, it helps track epigenetic markers associated with environmental exposures or lifestyle factors, giving researchers the ability to study interactions between genetics and environment on a molecular level.
ATAC-seq also complements other sequencing methods, such as ChIP-seq, by offering a broader view of regulatory regions, including promoters and enhancers. These data help build models for gene expression regulation, especially in understanding the dynamic changes in chromatin landscapes under various conditions.
Service you may intersted in
Chromatin remodeling plays a critical role in cancer development, neurological disorders, and autoimmune diseases. ATAC-seq is particularly useful in identifying mutations and abnormal regulatory elements associated with these diseases.
Cancer is driven not only by genetic mutations but also by epigenetic alterations, which influence gene expression patterns and cellular behavior. ATAC-seq provides an effective approach for studying these epigenetic modifications by mapping accessible chromatin regions across different cancer types and stages4. It helps researchers discover aberrant enhancer and promoter activities responsible for oncogene activation and tumor suppressor silencing.
ATAC-seq is crucial in pinpointing non-coding mutations that impact chromatin accessibility, which can lead to tumor progression. For instance, in a large-scale study using ATAC-seq data from 410 tumor samples, researchers identified over 562,709 transposase-accessible DNA elements, including distal enhancers that are linked to cancer progression through chromatin accessibility changes5.
Single-cell ATAC-seq (scATAC-seq) helps dissect tumor heterogeneity, revealing distinct subpopulations of cancer cells. In acute lymphoblastic leukemia (ALL), scATAC-seq has been used to uncover chromatin accessibility differences between subclones, which allows for better understanding of how different populations within the same tumor contribute to treatment response and progression6.
ATAC-seq plays a role in studying how cancer cells acquire resistance to therapies. For example, it was shown that chromatin regulators contribute to drug resistance by altering transcription factor binding in resistant cells, which can be tracked by comparing ATAC-seq profiles before and after treatment. These findings enable the development of combination therapies aimed at overcoming resistance7.
ATAC-seq helps profile the immune cells within the tumor microenvironment. By identifying changes in regulatory elements of immune cells, such as T-cells, ATAC-seq provides insights into how the immune system interacts with tumors, which is essential for developing immune checkpoint inhibitors. A recent study linked transcription factor activity in immune cells to specific immune responses within tumors8.
Neurodevelopmental and neurodegenerative diseases often involve complex interactions between genetics and the epigenome. ATAC-seq allows for the investigation of how chromatin accessibility changes contribute to conditions such as autism spectrum disorders (ASD), schizophrenia, and Alzheimer's disease.
Here are specific examples for each point regarding the use of ATAC-seq in neurodevelopmental and neurodegenerative diseases:
ATAC-seq has been applied to study chromatin accessibility during brain development, helping to uncover mutations in regulatory elements. For instance, a study on *Activity-Dependent Neuroprotective Protein (ADNP)* identified chromatin remodeling as a key factor in the development of ASD, revealing how mutations in ADNP affect neural tube formation and synaptic development, predisposing individuals to ASD9.
In schizophrenia, ATAC-seq has been utilized to investigate chromatin accessibility differences in neurons and glial cells. One study revealed that regions associated with schizophrenia exhibited significant differences in chromatin accessibility when compared to healthy controls, providing insights into disease-associated regulatory elements and the disrupted gene networks involved in psychiatric conditions10.
ATAC-seq has provided valuable data on chromatin changes in postmortem brain tissues from patients with Alzheimer's disease. Researchers have identified epigenetic signatures related to synaptic plasticity, inflammation, and protein aggregation, which are crucial in the pathology of Alzheimer's. For example, single-cell transcriptomics combined with ATAC-seq helped identify cell-type-specific changes in chromatin accessibility, advancing our understanding of how Alzheimer's progresses at the molecular level10.
The ability of ATAC-seq to uncover epigenetic changes in brain tissue enables new avenues for therapeutic development, including gene therapies and epigenetic drugs. Additionally, scATAC-seq is valuable for studying cell-type-specific chromatin dynamics in mixed populations of neurons, astrocytes, and microglia, which are often implicated in neuroinflammation and neurodegeneration.
In autoimmune diseases, the immune system mistakenly attacks healthy tissues, often driven by dysregulation in immune cell gene expression. ATAC-seq helps identify how chromatin changes in immune cells contribute to autoimmune responses, offering new insights into the molecular mechanisms underlying these disorders11.
ATAC-seq has revealed significant chromatin accessibility changes in T and B cells, particularly in autoimmune diseases like systemic lupus erythematosus (SLE). One study showed that in SLE, B cells exhibited altered chromatin landscapes that regulated cytokine expression, such as IL-6 and TNF-α, contributing to the abnormal activation of immune cells12.
In diseases like rheumatoid arthritis (RA) and multiple sclerosis (MS), ATAC-seq has identified specific epigenetic biomarkers in immune cells, such as B and T cells. These biomarkers are predictive of disease onset and progression. Additionally, they are useful in tracking the effectiveness of treatments over time, helping in therapeutic monitoring13.
ATAC-seq has helped map regulatory elements such as enhancers and promoters controlling inflammatory genes like IL-8, IL-6, and TNF-α. These elements are frequently dysregulated in autoimmune conditions like RA and lupus, driving chronic inflammation12. Understanding these epigenetic alterations provides insights into the mechanisms that underlie persistent immune activation.
ATAC-seq has become a key tool in identifying therapeutic targets in autoimmune diseases. Drugs targeting epigenetic enzymes, such as histone deacetylase inhibitors (HDAC inhibitors), are under investigation for modulating chromatin states and suppressing aberrant immune responses. ATAC-seq also assists in repurposing existing drugs by identifying shared regulatory elements between different autoimmune diseases, guiding the development of broad-spectrum treatments.
ATAC-seq is increasingly employed in drug discovery to identify epigenetic targets and evaluate drug efficacy. The chromatin landscape provides essential information about the state of gene expression in different conditions, offering insight into how potential drugs might alter gene activity.
In preclinical drug testing, ATAC-seq can determine how compounds affect chromatin accessibility across various tissues, helping to predict side effects or off-target effects. For example:
Furthermore, ATAC-seq is useful in drug repurposing efforts by comparing the chromatin accessibility profiles induced by different drugs, identifying new applications for existing drugs14.
ATAC-seq plays a crucial role in studying the dynamics of chromatin accessibility during development. In embryogenesis, tissues undergo multiple stages of differentiation, with each stage marked by changes in chromatin accessibility. ATAC-seq enables researchers to:
ATAC-seq has been used to study how chromatin accessibility changes during different stages of embryogenesis. For example, studies using ATAC-seq have shown how chromatin regions open and close in a regulated sequence to control gene expression during the development of tissues like the neural crest and mesoderm in vertebrates15.
ATAC-seq helps in identifying master regulators and transcription factors that guide cellular differentiation. In neural progenitor cells (NPCs), for instance, ATAC-seq identified key transcription factors that control the differentiation process, which could be linked to broader developmental pathways in embryogenesis.
ATAC-seq is used to investigate how chromatin accessibility patterns differ in healthy and diseased development, particularly in congenital disorders. For instance, in congenital heart diseases, ATAC-seq has been applied to reveal how accessibility changes in regulatory regions may lead to developmental anomalies in the heart.
The technique has also shed light on reprogramming processes, such as the conversion of somatic cells to induced pluripotent stem cells (iPSCs)16. ATAC-seq provides insights into how the epigenetic landscape changes during reprogramming and how it affects the maintenance of stem cell identity.
Single-cell ATAC-seq (scATAC-seq) offers a groundbreaking approach to exploring the diversity and complexity of chromatin accessibility at the individual cell level. It plays a crucial role in understanding cellular heterogeneity, a fundamental feature of many biological processes and disease states17. Unlike bulk ATAC-seq, which provides an average chromatin accessibility profile for a population of cells, scATAC-seq resolves these profiles for individual cells, revealing subtle differences that contribute to tissue function, development, and disease.
Service you may intersted in
In tissues with complex cellular composition, such as the brain, immune system, or tumors, scATAC-seq is essential for identifying previously uncharacterized cell types and subpopulations. For instance:
This level of resolution allows researchers to study cell states more precisely, including dynamic transitions such as those occurring during differentiation, proliferation, or stress responses.
One of the powerful applications of scATAC-seq is the reconstruction of lineage trajectories. By tracking changes in chromatin accessibility, researchers can infer how stem cells and progenitors transition into mature cell types. For example:
These trajectory analyses offer valuable insights into the regulatory landscapes governing tissue development, regeneration, and repair.
scATAC-seq is instrumental in revealing disease-associated epigenetic changes within specific cell populations. This is particularly relevant in diseases with a heterogeneous cellular landscape, such as cancer and autoimmune disorders.
scATAC-seq data is often integrated with other single-cell omics methods, such as single-cell RNA-seq (scRNA-seq), to correlate chromatin accessibility with gene expression. This multi-omics approach provides a more comprehensive understanding of gene regulation and cell function, uncovering the relationships between chromatin landscapes, transcriptional profiles, and phenotypic diversity.
References
Terms & Conditions Privacy Policy Copyright © CD Genomics. All rights reserved.