We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

Tumor immunotherapy has become a focal point in cancer treatment, with T cell immunotherapy holding great promise. TCR-T and CAR-T therapies emerged contemporaneously; however, TCR-T technology has been hampered by difficulties in T cell acquisition and purification, target selection, and HLA allele matching. Nevertheless, recent research endeavors have been dedicated to surmounting these obstacles. This study explores the utilization of TCR sequencing in CAR-T therapy and its implications for personalized cancer treatment. Here shows the application of TCR sequencing in CAR-T therapy for tumor.
In this investigation, researchers conducted comprehensive analyses of T cells in tumor and adjacent tissues from lung cancer patients. Through single-cell mRNA sequencing (scRNA-Seq) and TCR sequencing (TCR-Seq), they aimed to dissect the characteristics of tumor-infiltrating T cells. CD45+ CD3+T cells were isolated from tumor, peritumor, normal tissues, and peripheral blood of five untreated lung cancer patients. Subsequently, scRNA-Seq and TCR-Seq were performed on the 10x genomics platform. After data processing and batch effect correction, T cells were classified into CD4+ and CD8+ subsets, and further clustered for detailed analysis.
Take the Next Step: Explore Related Services
Learn More
By developing a more effective Tas cell sorting method, combined with single cell transcriptome analysis, TCR sequencing and new antigen stimulation, Tas cells and their biomarkers can be identified, so as to further develop personalized TCR-T cell therapy and improve the effectiveness and success rate of tumor treatment.
In vitro tumor antigen stimulation system
To explore antigen specificity, an in vitro tumor antigen stimulation system was established. TCR sequences were constructed and transfected into peripheral blood mononuclear cell (PBMC)-derived T cells to generate TCR-T cells. Tumor and surrounding tissue samples underwent whole exome sequencing (WES) and RNA sequencing (RNA-Seq) to identify potential tumor-specific mutations. B lymphocytes transformed by EB virus infection were utilized as antigen-presenting cells (APCs). TCR-T cells were co-cultured with APCs transfected with tandem genes containing tumor mutation sequences, and the interferon-gamma (IFN-γ) production was measured to assess antigen response. Additionally, patient tumors were transplanted into immunodeficient NOD-SCID IL-2 receptor γ-deficient (NSG) mice to establish patient-derived xenograft (PDX) models for in vivo therapeutic experiments.
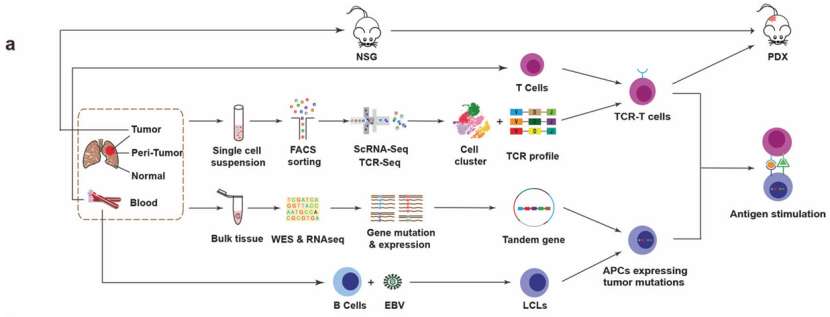 Scheme of overall study design(He et al., 2022)
Scheme of overall study design(He et al., 2022)
Identification of tumor-rich T cell clusters
In five untreated lung cancer patients, CD45+ CD3+T cells were isolated from tumor, tumor surrounding tissues, normal tissues and peripheral blood, and scRNA-seq and TCR-seq were performed on 10x genomics platform to track T cell clones in tumor and its adjacent tissues.
The cells with low quality and double clearance were screened out, and 178 million RNA transcripts of 42,511 T cells with CDR3 sequences were obtained, and 22,592 unique TCR were identified. After the batch effect was corrected by Seurat, T cells were divided into CD4+and CD8+T cells, and 10 CD4 and 9 CD8 clusters were identified by cluster analysis for further analysis.
Amplified T cells were detected in the subset of T cells. Further analysis of CD4+and CD8+T cells revealed multiple clusters, including the traditional naive, effector, memory and depletion clusters and CD4+regulatory T cells (Tregs). At the same time, two clusters with active mitosis were also identified in CD4+and CD8+T cells, which were characterized by high expression of MKI67, TK1 and STMN1.
Examining the distribution of each cluster in the tissue, it was found that some T cell clusters were enriched in the tumor, such as depleted clusters, CD4+ Tregs and clusters with active mitosis. CD4-C9-MKI67 cluster was also found in normal tissues. Highly expressed depletion markers are not limited to two depletion clusters, but CD4+Tregs and CD8 cluster with active mitosis also show similar gene expression levels. Moderate depletion genes were also observed in CD4+ clusters with active mitosis. These results show that T cell clusters rich in tumors have the characteristics of fatigue gene expression, which indicates that T cells in these clusters have experienced antigen stimulation.
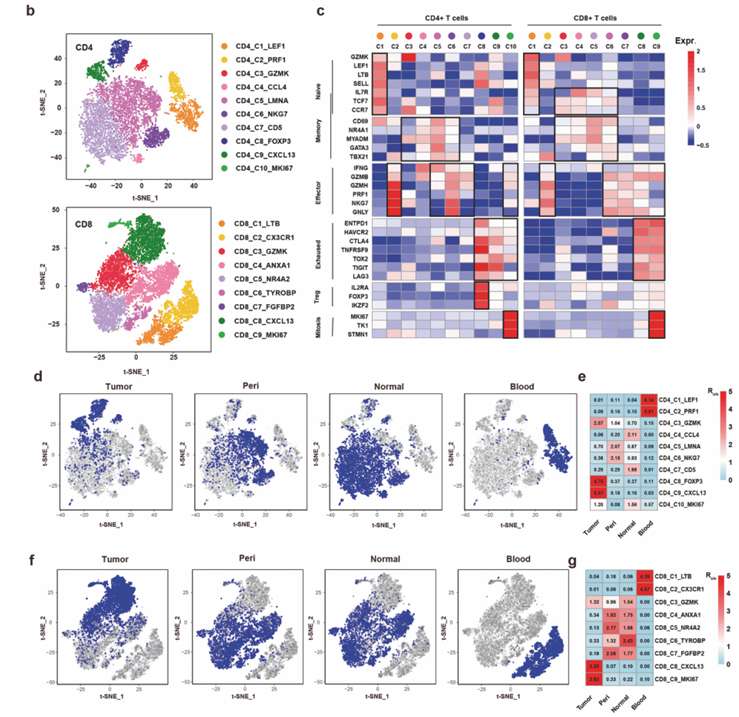 ScRNA-seq identified tumor-enriched T cell clusters (He et al., 2022)
ScRNA-seq identified tumor-enriched T cell clusters (He et al., 2022)
Take the Next Step: Explore Related Services
Tumor-specific clonal expansion
In order to identify the biomarkers of tumor-specific T cells, the researchers compared the gene expression of T cells with tumor-specific TCR with T cells with tissue-sharing TCR in tumor CD4+ or CD8+ T cells. In addition, CD4+ Treg cells were compared with CD4+ T cells with tissue-sharing TCR. By comparing these T cell populations, the researchers identified many differentially expressed genes.
The researchers selected tumor and surrounding tissue samples from patients with lung cancer for full exon sequencing, detected tumor-specific mutations, and used batch RNA-seq to evaluate the expression of each mutation. In order to increase the sample size, tumor samples from five other lung cancer patients were also used for single cell RNA-seq. The TMB of 10 patients was calculated as the average number of mutations per megabase.
The frequency of T cell cloning in tumor and its adjacent tissues is calculated to identify people with tumor-specific clonal expansion. The distribution of tumor TCR in tumor and its adjacent tissues was compared, and tissue sharing and tumor-specific TCR were found. In tumors, the cloning frequency of CD8+T cells is higher than that of CD4+T cells, indicating that tumor antigens are mainly presented by MHC class I molecules in these patients.
Tumor-specific TCR dilatation exists in tumor-rich clusters (C9 and C10 in CD4+ T cells and C8 and C9 in CD8+ T cells), which supports the hypothesis that tumor-specific TCR dilatation is mainly caused by the stimulation of new antigens from tumor mutation.
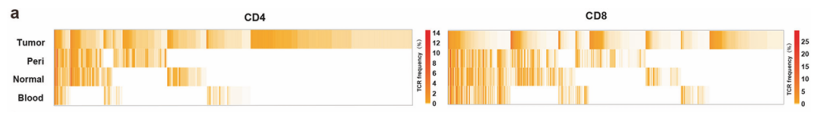 Heatmap showing the frequency of each TCR in the four tissues (He et al., 2022)
Heatmap showing the frequency of each TCR in the four tissues (He et al., 2022)
The Proportion of Tumor-specific TCR
By measuring the gene expression in the specifically dilated TCR, the biomarkers of T cells that recognize tumor antigens were detected. For tumor CD4+ or CD8+ T cells, the TCR with specific expansion was compared with that with tissue sharing, and CD4+ Treg cells were also compared with CD4+ T cells with tissue sharing TCR. In addition, the expression of CXCL13 was examined for each T cell cluster. Subsequently, the first 20 tumor TCR in each tumor were selected and their distribution in tumor and adjacent tissues was compared. Finally, the frequencies of four types of TCR in CD4+CXCL13+,CD4+CXCL13-, CD8+CXCL13+and CD8+CXCL13- T cells in 10 patients were compared.
Exon sequencing was performed to detect tumor-specific mutations, and batch RNA-seq was performed to evaluate the expression of each mutation. Single cell RNA sequencing was performed on tumor samples of five lung cancer patients to increase the sample size. TMB was calculated to represent the new antigen load, and T cells from 10 tumor samples were allocated into 10 CD4 and 9 CD8 clusters by using the R software package "cellassign". The proportion of each cluster in tumor CD4+or CD8+T cells was calculated.
In C9 and C10 of CD4+T cells and C8 and C9 of CD8+T cells, the proportion of clusters with tumor-specific TCR was positively correlated with TMB, while most other clusters were negatively correlated with TMB, but there was no statistical significance.
Immune stimulation of new antigen in tumor is the main cause of T cell expansion, and the percentage of CXCL13+T cells in tumor is positively correlated with TMB, which proves that CXCL13 is a biomarker of tumor-specific T cells.
CXCL13 as a biomarker
In order to identify the biomarkers of TCR with specific expansion in tumor, the researchers compared the gene expression of T cells with tumor-specific TCR and T cells with tissue-sharing TCR in tumor CD4+or CD8+T cells. CD4+Treg cells were also compared with CD4+T cells with tissue-sharing TCR.
Many differentially expressed genes were identified in these T cell populations, among which CXCL13 was specifically expressed in CD4+and CD8+T cells with tumor-specific TCR. We further examined the expression of CXCL13 in each T cell cluster, and found that tumor-rich clusters (C9, C10 in CD4+T cells and C8, C9 in CD8+T cells) expressed high levels of CXCL13. We selected the top 20 tumor TCR in each CD4+CXCL13+,CD4+CXCL13-, CD8+CXCL13+and CD8+CXCL13-T cell, and compared their distribution in the tumor and its adjacent tissues. Tumor-specific TCR amplification was observed in CXCL13+T cells, while TCR in CXCL13- T cells was shared in tissues. We also compared the frequencies of four TCR(1α1β, 1α2β, 2α1β and 2α2β) in CD4+CXCL13+,CD4+CXCL13-, CD8+CXCL13+and CD8+CXCL13- T cells of 10 sequencing patients, and found their CXCL13+and CXCL in CD4+or CD8+T cells.
In 10 NSCLC patients, the percentage of CXCL13+cells in total tumor T cells was positively correlated with TMB. The analysis of tumor RNA-seq data from the cancer genome map (TCGA) database also showed that the median expression of CXCL13 was significantly correlated with TMB in various cancer types.
CXCL13 is a unique marker of tumor-specific expanded T cells.
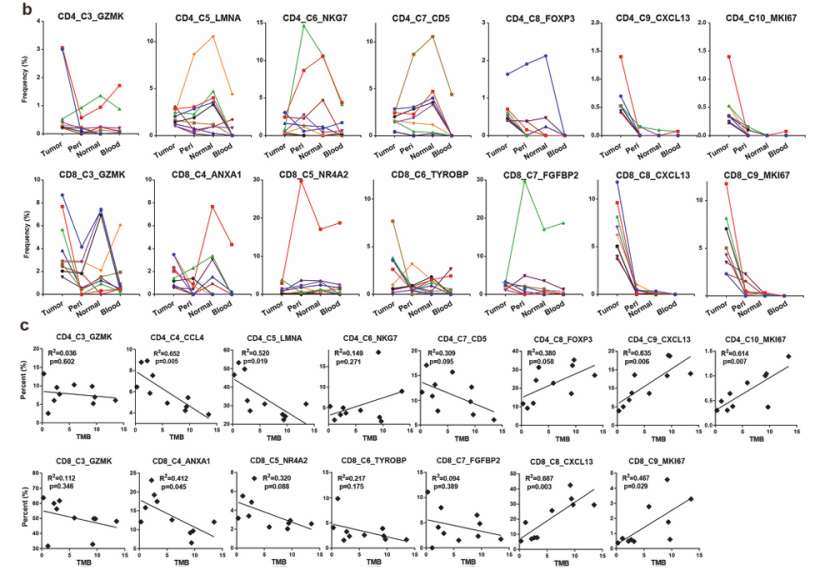 Tumor-enriched T cells were expanded by tumor antigens (He et al., 2022)
Tumor-enriched T cells were expanded by tumor antigens (He et al., 2022)
Tumor-specific TCR for new antigen stimulation
The researchers determined whether tumor-specific TCR recognized tumor-specific antigen (TSA) through tumor antigen stimulation experiment in vitro.
Using tumor samples from P4 and P5 patients, the first five TCRs were selected from CD4+CXCL13+, CD4+CXCL13, CD8+CXCL13+ and CD8+CXCL13− T cells. These TCRs were synthesized and constructed into lentiviral vectors expressing surface marker Thy1.1. These TCRs were transduced into allogenic T cells collected from peripheral blood mononuclear cells (PBMCs), and 20 TCR-T cell lines were generated for each patient. 100 mutation sites with high to medium mRNA level were selected from each patient's tumor as potential tumor antigens, and these sites were detected by WES and bulk RNA-seq. Five tandem genes were constructed and transduced into allogeneic lymphoblastoma cell lines (LCLs).
Each TCR-T cell line was co-cultured with mixed LCLs transduced with tandem genes, and untransfected LCLs were used as control. The IFNγ production in TCR-T cell line was measured by intracellular staining method to evaluate the response to antigen stimulation. TCR-T cells producing at least 3 times of IFNγ in the control group were determined to be positive. The experimental results showed that in P4 patients, one CD4+ and four CD8+ tumor-specific TCR responded positively to tumor antigen stimulation, but no positive response was detected in tumor-sharing TCR. In P5 patients, one CD4+ and all CD8+ tumor-specific TCR responded positively to tumor antigen stimulation, but no positive response was detected in tumor-sharing TCR. These data directly prove that tumor-specific TCRs recognize tumor antigens.
Based on the above experimental results, the researchers concluded that CXCL13+ T cells are Tas cells in tumors, and tumor-specific TCRs can recognize tumor antigens and have the potential to treat autologous tumors.
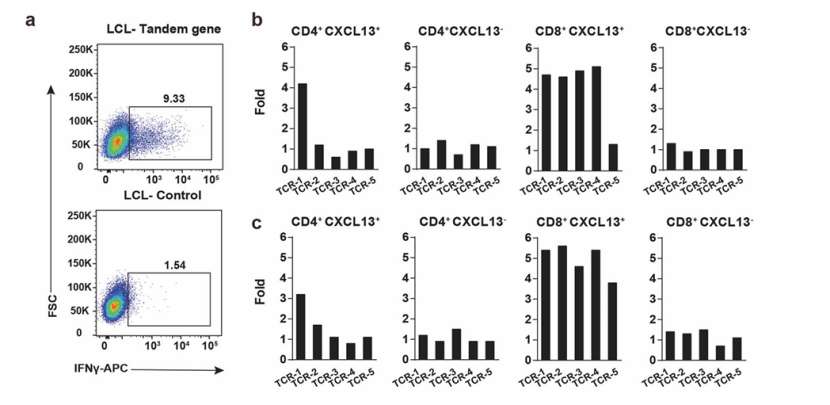 FACS plots showing the IFNγ expression in TCR-T cells (He et al., 2022)
FACS plots showing the IFNγ expression in TCR-T cells (He et al., 2022)
TCR-T Cells transformed with TCR of Tas cells
The purpose of the researchers is to study whether TCR-T cells modified by TATCRS can treat autologous tumors.
Researchers used tumor samples from P4 or P5 patients to establish a PDX tumor model in NSG mice. Then, the researchers used the first three TCR selected from CD4+CXCL13+,CD4+CXCL13, CD8+CXCL13+and CD8+CXCL13− T cells in each tumor tissue to transform TCR-T cells, and used untransfected T cells as the control group. Then these modified TCR-T cells were transplanted into mice for treatment.
The researchers found that TCR-T cells expressing TCR from CD8+CXCL13+T cells have obvious therapeutic effect on autologous PDX tumor in P4 or P5 patients. For TCR-T cells transformed with TCRs from CD4+CXCL13+T cells, only TCRs which showed positive response to new antigen stimulation in vitro showed therapeutic effect. No therapeutic effect was observed in the treatment of T cells modified by TCRs from CXCL13− T cells.
These data show that the TCR-T cells of the modified TATCRS can effectively treat autologous tumors.
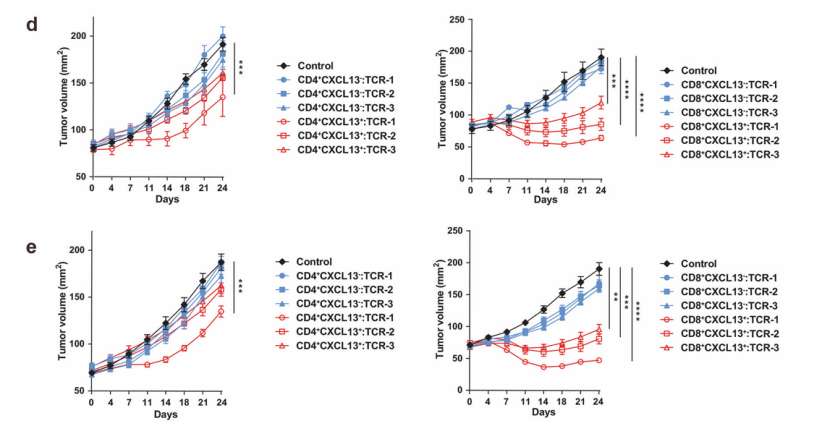 T cells from tumor-enriched clusters recognize neoantigens (He et al., 2022)
T cells from tumor-enriched clusters recognize neoantigens (He et al., 2022)
Predictive value of CXCL13 for ICB response
To study whether the expression level of CXCL13 can predict the response of immune checkpoint inhibitor (ICB) treatment.
Objective response rate (ORRs) of ICB in reported clinical trials was collected, and the expression of CXCL13 in different cancer types was analyzed. At the same time, the expression of CXCL13 in tumor was detected by RNAseq and immunohistochemistry (IHC), and the relationship between overall survival rate (OS), progression-free survival rate (PFS) and ICB response related to ICB treatment was evaluated.
The median expression level of CXCL13 is significantly correlated with ORRs of ICB in various cancer types. High CXCL13 expression is related to better OS and better response to ICB treatment in tumor patients. IHC showed that CXCL13 was specifically expressed in CD4+ and CD8+ T cells, and the expression was higher in tumors with TLS. High level of CXCL13 protein is related to patients' better PFS.
The level of CXCL13 can accurately predict the response of tumor patients to ICB treatment, and the expression of CXCL13 may be used as an effective biomarker for ICB treatment.
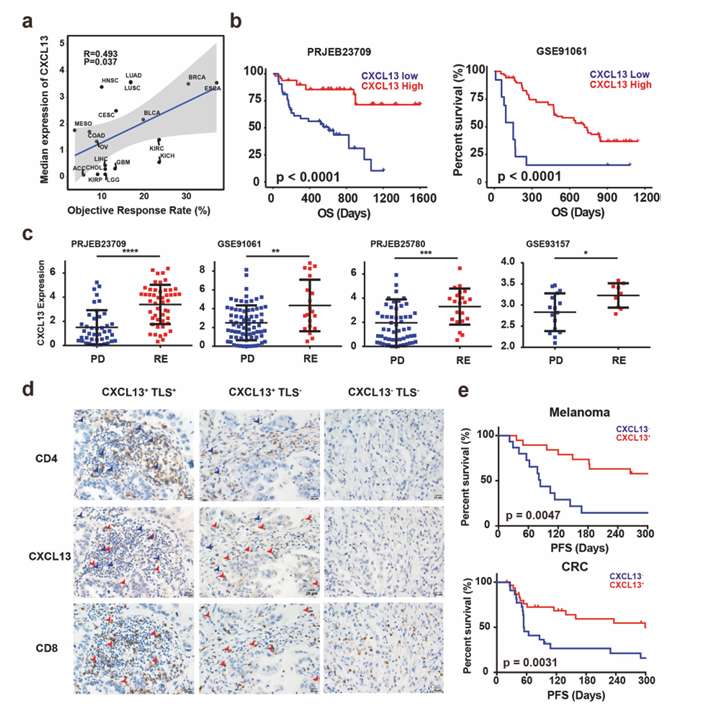 Expression of CXCL13 predicts response to ICB (He et al., 2022)
Expression of CXCL13 predicts response to ICB (He et al., 2022)
Identification of Tas cell surface markers
In this study, the researchers identified and studied an important class of anti-tumor immune cells-TAS cells by analyzing the expression level of CXCL13 in tumor infiltrating lymphocytes. Then, the researchers selected surface markers highly expressed in CD4+ and CD8+ Tas cells to promote the sorting and identification of Tas cells. Finally, the researchers tested whether these markers can be used to sort Tas cells in tumors by flow cytometry and whether they respond to autologous tumors.
The researchers selected CD200 and ENTPD1 as surface markers. CD4+and CD8+Tas cells were separated from tumor samples of different cancer patients by flow cytometry, and the expression level of CXCL13 in them was evaluated by qPCR. The researchers also evaluated the response of CD4+CD200+ and CD8+ ENTPD1+Tas cells to autologous tumor cells in vitro.
The researchers found that CD200 and ENTPD1 can be used to sort CD4+and CD8+Tas cells, and the expression level of CXCL13 in these Tas cells is significantly higher than that of the corresponding negative T cells. At the same time, autologous tumor cells can activate the immune response of CD4+CD200+ and CD8+ ENTPD1+Tas cells.
CD200 and ENTPD1 are surface markers that can be used to identify and sort CD4+and CD8+Tas cells with tumor infiltration. The surface markers of Tas cells can be used to sort Tas cells in tumors by flow cytometry, which is helpful to deeply understand the functions and regulatory mechanisms of these important anti-tumor immune cells.
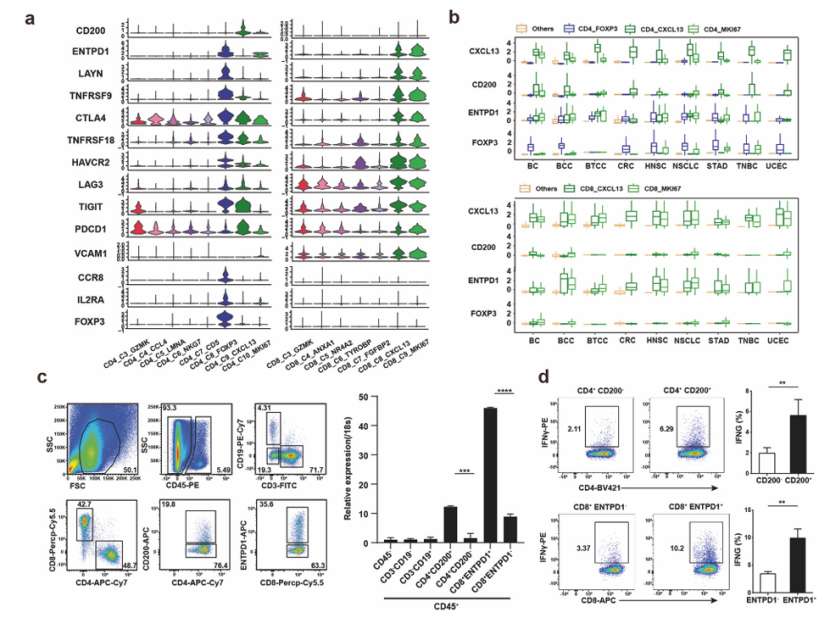 Identification of surface markers for Tas cells (He et al., 2022)
Identification of surface markers for Tas cells (He et al., 2022)
In summary, the application of TCR sequencing in CAR-T therapy represents a promising avenue for advancing cancer treatment, and continued research efforts are warranted to fully exploit its potential.
Reference

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy