We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

Phenotypic analysis has always been the key to understand gene function and cell process. Everyone must be familiar with the 2020 Nobel Prize in Chemistry "CRISPR". The gene screening technology based on CRISPR is a powerful research method of functional genomics. The pooling and array screening of CRISPR have influenced and changed biological research and accelerated its development, which is favored by many researchers. CRISPR can be used to identify gene functions, clarify molecular pathways, discover potential therapeutic targets, etc. However, this technology relies on batch analysis, which may ignore complex molecular mechanisms and cell type-specific functions, and reading depends on the measurement of different phenotypes. If cell groups show heterogeneity in gene expression and response, especially in cancer cell lines showing different drug resistance or immune response, these changes may be ignored.
Traditional gene editing methods can only observe the average response of the whole cell population, but single cell CRISPR technology (scCRISPR) which combines CRISPR screening sequencing with single cell sequencing allows different gene editing for thousands of cells at the same time, and then observe their responses one by one, so as to study the gene function and cell behavior at the single cell level with high resolution, and realize high-content phenotype analysis and Qualcomm-quantity gene screening. scCRISPR can be applied to the research of transcription analysis, chromatin accessibility, epigenetics and single cell imaging, such as accurately identifying key genes and their functions, revealing the interaction between genes, exploring the roots of cell heterogeneity, and studying the determinants of cell fate in complex biological processes.
scCRISPR has developed rapidly in recent years, and researchers have systematically reviewed the research progress of scCRISPR. According to its sgRNA identification method and the combined single cell analysis platform, scCRISPR methods are mainly divided into the following four categories.
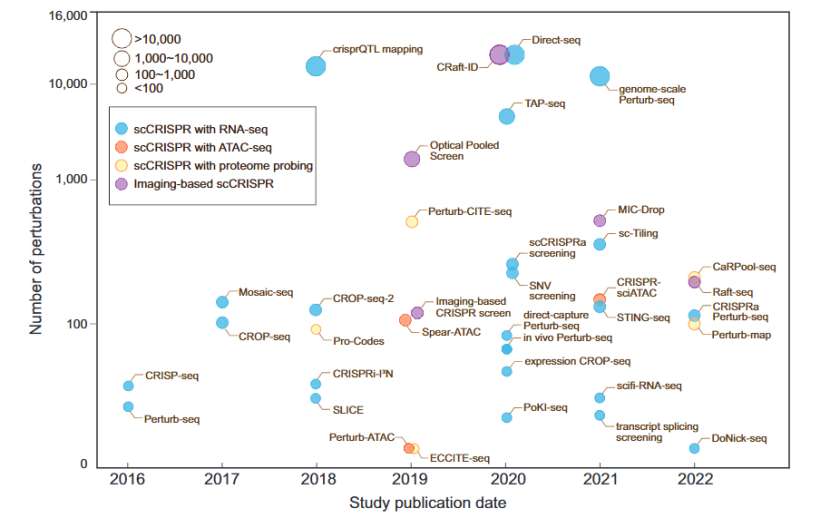 Progress of scCRISPR method (Chen et al., 2023)
Progress of scCRISPR method (Chen et al., 2023)
The core of scCRISPR method is how to trace back sgRNA information effectively after single cell analysis, and associate phenotype with corresponding gene disturbance. Let's take a look at the brief introduction of these four kinds of scCRISPR methods:
scCRISPR combined with RNA-seq
It relates sgRNA information with transcriptome information, and studies the influence of gene disturbance by analyzing the gene expression profile of a single cell.
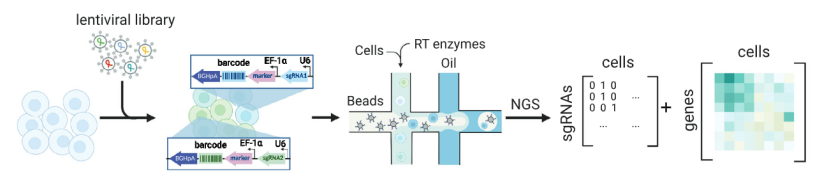 scCRISPR with RNA-seq (Atray et al., 2016)
scCRISPR with RNA-seq (Atray et al., 2016)
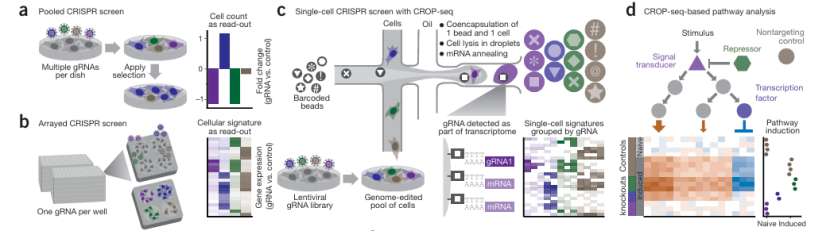 CROP-seq enables pooled CRISPR screening with single-cell transcriptome readout (Paul et al., 2017)
CROP-seq enables pooled CRISPR screening with single-cell transcriptome readout (Paul et al., 2017)
scCRISPR combined with ATAC-seq
This method correlates sgRNA information with chromatin accessibility information, study the influence of gene disturbance on chromatin opening state, and then explore the gene regulation mechanism.
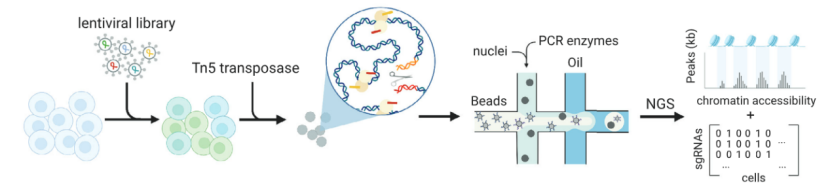 scCRISPR with ATAC-seq (Atray et al., 2016)
scCRISPR with ATAC-seq (Atray et al., 2016)
scCRISPR combined with protein group detection
It is a method to correlate sgRNA information with protein group information, to study the influence of gene disturbance on protein expression, and then to explore the gene function and its influence on cell signal pathway.
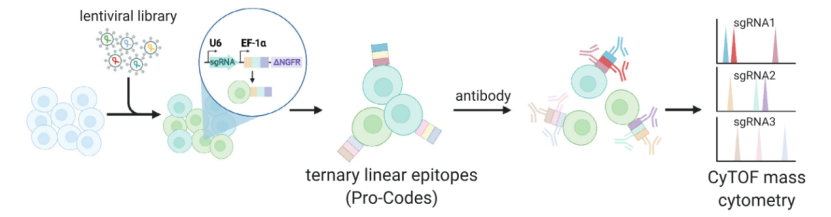 scCRISPR with proteome probing (Atray et al., 2016)
scCRISPR with proteome probing (Atray et al., 2016)
Imaging-based scCRISPR
It captures the phenotypic changes of cells by imaging technology, and determines the sgRNA information by in-situ sequencing or other methods, and spatially associates genotypes with phenotypes.
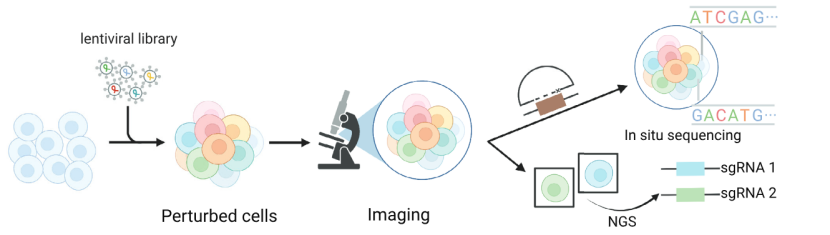 Imaging-based scCRISPR (Atray et al., 2016)
Imaging-based scCRISPR (Atray et al., 2016)
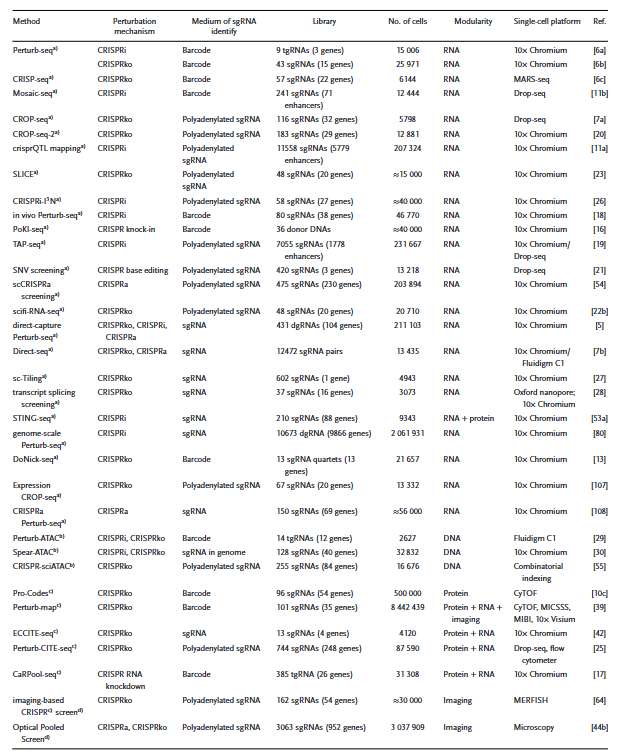 Overview of the scCRISPR methods (Cheng et al., 2023)
Overview of the scCRISPR methods (Cheng et al., 2023)
Take the Next Step: Explore Related Services
Learn More
The application prospect of scCRISPR technology is still very broad. This technology can not only analyze the regulatory network in complex biological systems with high resolution, but also accurately correlate genotypes and phenotypes at the single cell level, providing a new perspective for in-depth understanding of cell heterogeneity and gene function.
Precision medicine
scCRISPR technology of precision medicine can be used to screen and identify gene mutations related to diseases, evaluate the efficacy of drugs on different cell subsets, and provide basis for the formulation of personalized medical programs. For example, scCRISPR technology is used to screen targeted drugs for tumor-specific mutations, or to evaluate the effects of immunotherapy drugs on different immune cell subsets. Researchers used SLICE technology (sgRNA lentivirus infection combined with Cas9 protein electroporation system) to combine CROP-seq with electroporation delivery of CRISPR reagent to study the immune response of human primary T cells to solid tumors. They found that compared with the traditional lentivirus transduction, electroporation delivery of CRISPR reagent can significantly improve the efficiency of gene editing, and can complete gene editing in a short time, which is suitable for studying cell types with short life cycle.
Scholars developed Perturb-CITE-seq method by combining Perturb-seq and Cite-Seq technology, which was used to simultaneously analyze sgRNA, transcriptome and cell surface proteins of melanoma cells. They used this method to study the drug resistance mechanism of melanoma cells to immunotherapy drugs, and found that the expression levels of immune checkpoint proteins such as PD-1 and CTLA-4 were closely related to the drug resistance of tumor cells.
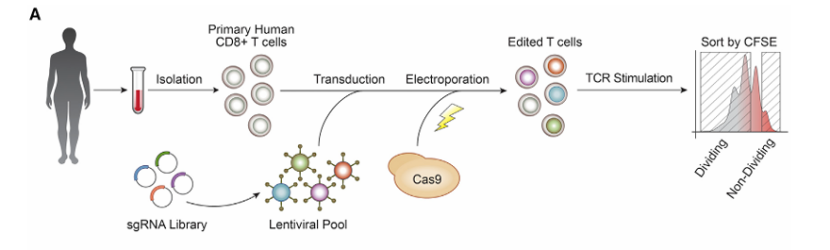 Diagram of a hybrid system of sgRNA lentiviral infection and Cas9 electroporation (Eric et al., 2018)
Diagram of a hybrid system of sgRNA lentiviral infection and Cas9 electroporation (Eric et al., 2018)
Drug research and development
Drug research and development can be used to screen drug targets by Qualcomm quantity, evaluate the effect of drugs on cell function, and accelerate the process of drug research and development. For example, scCRISPR technology is used to screen new targets of anti-tumor drugs, or to evaluate the effects of drugs on cell signaling pathways.
Researchers applied CRISPR technology to IPCS-derived neurons to study the genes related to neuron survival and function by using in (Isogenic, Integrated and Inducible Neurons, in system is a dCas9-KRAB expression IPCS cell line based on TALEN technology, which can knock down genes efficiently and differentiate into glutamatergic neurons through NGN2 induction). This system provides a new model and tool for studying nervous system diseases.
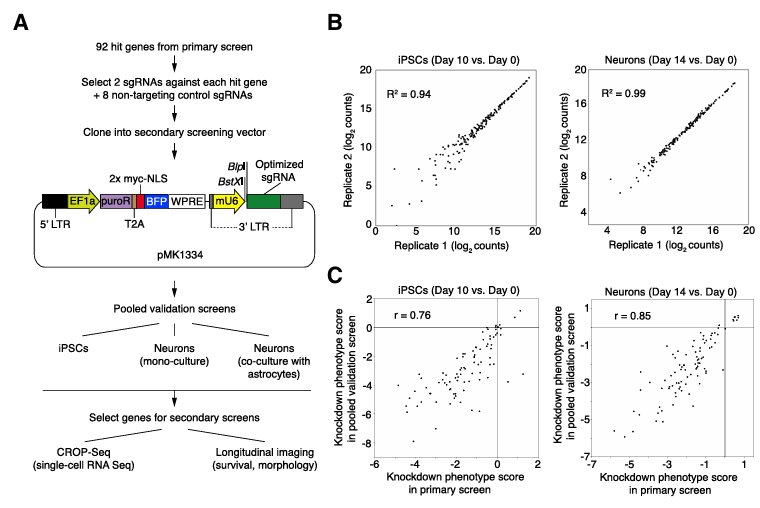 Strategy for validation of hit genes (Ruilin et al., 2019)
Strategy for validation of hit genes (Ruilin et al., 2019)
Gene therapy
Gene therapy can be used to develop safer and more efficient gene therapy strategies, such as gene editing of hematopoietic stem cells by scCRISPR technology to treat hereditary blood diseases.
Researchers used PoKI-seq technology to conduct large-scale gene knock-in screening of primary human T cells, in order to find chimeric antigen receptor (CAR) which can enhance the anti-tumor function of T cells. They introduced the synthetic genes of Cas9-gRNA ribonucleoprotein and TCR homologous arm containing unique barcode into T cells by electroporation, and screened CAR which can enhance the anti-tumor function of T cells under immunosuppression conditions.
Study of disease mechanism
The study of disease mechanism can be used to study the mechanism of complex diseases, such as the interaction of different cell subsets in tumor microenvironment by scCRISPR technology, or the mechanism of neuron death in neurodegenerative diseases.
Researchers studied the functions of genes related to autism spectrum disorder (ASD) and neurodevelopmental retardation (ND) during the development of mouse embryonic brain by Perturb-seq technique in vivo. They injected the dgRNA library targeting multiple neurodevelopmental risk genes into the lateral ventricle of mouse embryos, and analyzed the mouse brain cells after birth by scRNA-seq, revealing the role of ASD/ND risk genes in mouse brain development, and found gene co-regulation modules related to five different subgroups of neurons.
Synthetic biology
Synthetic biology can be used to construct and transform artificial life systems. For example, scCRISPR technology is used to transform microbial cells to produce specific biological products.
Researchers developed the MIC-Drop platform, which used a single needle to continuously inject microdroplets into thousands of zebrafish embryos, and could make phenotypic analysis through macroscopic observation.
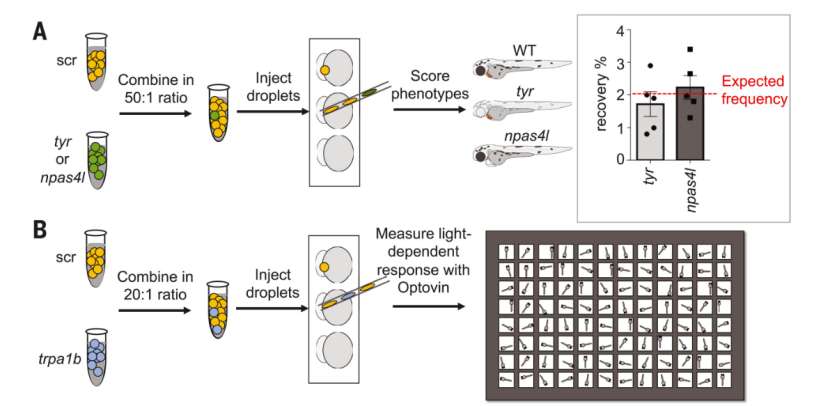 Schematic of a spike-in phenotypic (Saba et al., 2021)
Schematic of a spike-in phenotypic (Saba et al., 2021)
Future prospects
More efficient delivery system: At present, the application of scCRISPR technology is limited by delivery efficiency, especially for primary cells and in vivo research. Therefore, developing a more efficient and safer CRISPR delivery system is an important direction of scCRISPR technology development.
Expanding application scope: scCRISPR technology is mainly used to study gene function and regulatory network at present, and it can be extended to more fields (such as cell therapy, drug screening, disease model construction, etc.) in the future. For example, to build a more accurate disease model for studying the occurrence and development mechanism of diseases and screening therapeutic drugs.
Multiomics integration: By integrating scCRISPR technology with other single cell genomics technologies, the genotype-phenotype relationship of cells can be analyzed more comprehensively. For example, combining scCRISPR with single cell protein genomics, we can study the effect of gene knockout on protein expression, so as to understand the gene function more deeply.
Spatial transcriptomics: Combining scCRISPR technology with spatial transcriptomics, we can study the influence of gene disturbance on the spatial distribution of tissues. For example, to study the gene expression differences of different cell subsets in tumor microenvironment, so as to understand the mechanism of tumor occurrence and development more deeply.
In vivo study: Developing scCRISPR technology suitable for in vivo study will help to reveal the mechanism of gene function under physiological and pathological conditions. For example, study the effect of gene knockout on mouse model, so as to understand the role of gene function in the occurrence and development of diseases more deeply.
Main challenges
The off-target effect of sgRNA: The off-target effect of SGRNA is a big challenge of CRISPR technology, which may lead to unexpected gene editing and affect the accuracy of experimental results. In the future, it is necessary to develop more accurate sgRNA design methods and Cas enzymes to reduce the influence of off-target effect.
Cell type limitation: The application efficiency in some cell types is low, and in-situ editing is still difficult. It is necessary to optimize the vector system and develop tissue-specific Cas protein.
Data analysis: scCRISPR technology produces a huge amount of data, which has the characteristics of high dimension and sparsity. In the future, it is necessary to develop more powerful bioinformatics tools to analyze and interpret scCRISPR data.
Cost: It is necessary to develop more economical and efficient single-cell platform and CRISPR technology to reduce the application cost of scCRISPR technology.
Ethical issues: The application of scCRISPR technology involves gene editing, and relevant ethical norms need to be formulated.
References

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy