CRISPR Screening Sequencing
Overview of CRISPR screening sequencing
CRISPR/Cas9 system is a gene editor whose main advantage is its simplicity and versatility, which has been widely applied in engineering with gene editing. CRISPR/Cas9 system consists of type II Cas9 nuclease and single guide RNA (sgRNA). Cas9 and sgRNA form a complex that can recognize genes with PAM (protospacer adjacent motif) information. The system is based on the intracellular non-homologous end-joining mechanism and homologous recombination repair mechanism to repair the broken DNA, resulting in insertion or deletion of partial sequences of genes, and finally achieving targeted gene editing. In summary, CRISPR technology is applicable to almost any gene locus by directing Cas9 to make specific DNA modifications to target genes via sgRNA.
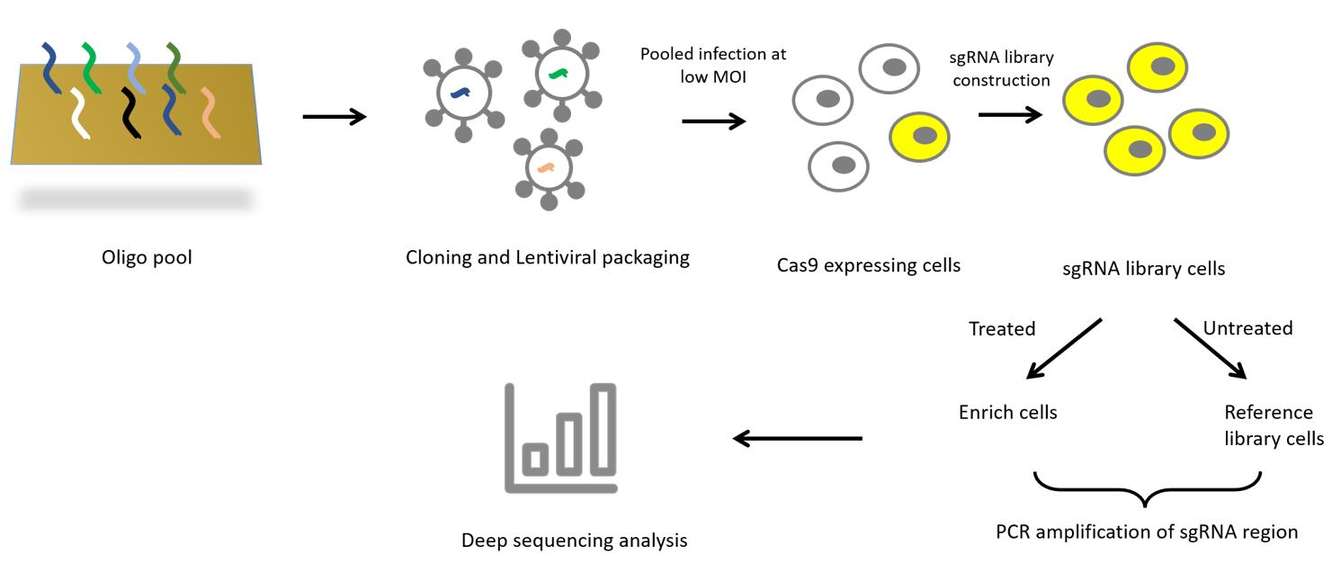 Figure 1. Schematic diagram of CRISPR screening
Figure 1. Schematic diagram of CRISPR screening
CRISPR/Cas9 technology enables genome-wide screening to identify and find genes and loci of interest. In order to achieve high-throughput CRISPR screening, it is first necessary to design genome-wide sgRNA libraries for the genes to be screened, infect the host cells at low infection complexes after lentiviral packaging, and test the changes in sgRNA abundance before and after screening under appropriate screening conditions to identify the candidate genes. CRISPR screening can identify genes affecting many physiological effects, such as drug resistance and drug sensitivity, making it a powerful tool in the biomedical and pharmaceutical fields. Based on this, CD Genomics offers you NGS-based high-throughput sequencing to support your research and projects related to efficient CRISPR screening.
The workflow of CRISPR screening sequencing service
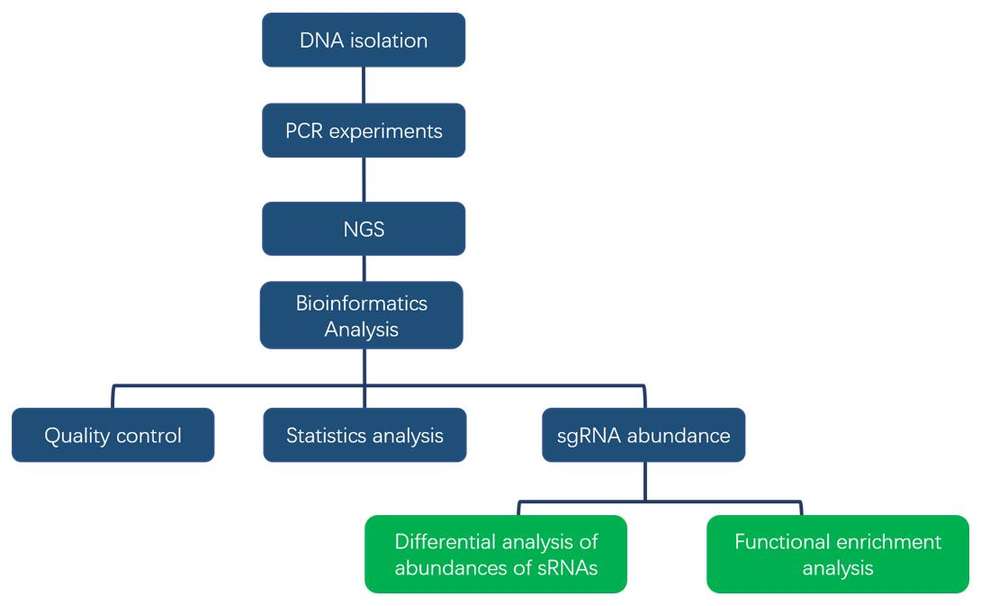
At CD Genomics, NGS-based high-throughput strategies can support your CRISPR screening process to achieve efficient and high-quality screening and analysis of targeted genes. The operational flow is mainly shown in the figure below, including sgRNA library design and synthesis, lentiviral vector construction and infection of host cells, phenotype selection, high-throughput sequencing and bioinformatics analysis.
01. Sequencing sample requirements
| Sample Type |
DNA Amount |
DNA Concentration |
Purity |
| Total DNA from all types of cells, blood, saliva, tissue, formalin-fixed samples, fresh frozen tissue samples, paraffin-embedded tissue, cancer liquid biopsy samples, colonies, swabs, etc. |
≥ 200 ng |
≥ 20 ng/μL |
OD260/280=1.8 - 2.0;
no degradation,
no contamination |
02. Library construction and NGS
NGS-based Illumina / MGI platform
03. Bioinformatics Analysis
- Reference alignment
- sgRNA abundance analysis
- CRISPR screening of target genes and phenotype relationship analysis
- Differential analysis of sgRNA abundance
Advantages of our CRISPR screening sequencing service profiling
- High-throughput NGS-based technology platform ensures short cycle times and accurate data
- Efficiently simplifies complex CRISPR screening process for high-throughput screening
- One-stop sequencing analysis, from the sample to be tested directly to the analysis report, greatly improving efficiency
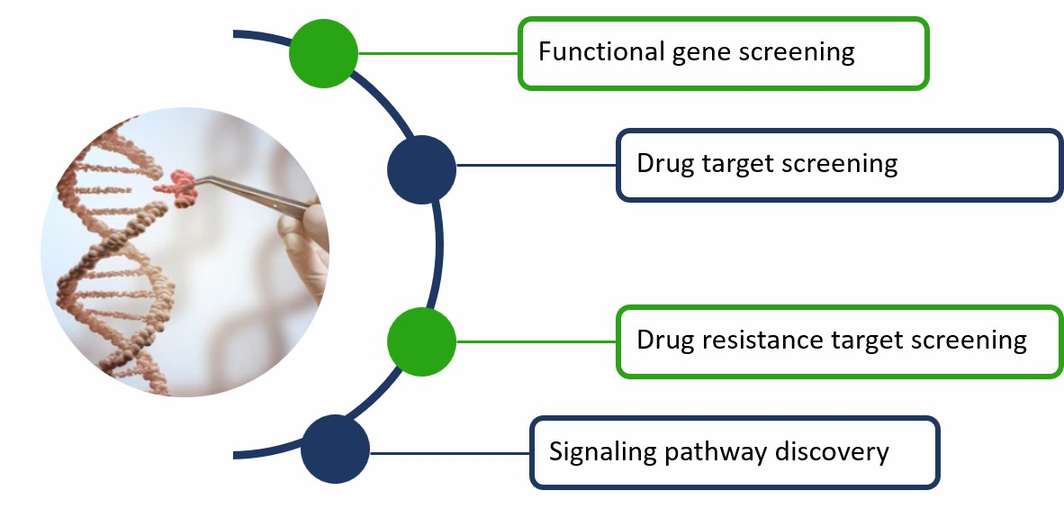
Applications in genome editing and engineering
CD Genomics provides NGS-based high-throughput sequencing services mainly for efficient CRISPR screening, which can be widely used for functional gene screening, drug target and resistance target screening, viral target screening and revealing signaling pathways, and gene interactions screening. CRISPR/Cas9 screening can not only screen key genes and non-coding DNA in pathological and physiological processes, but also reveal the molecular mechanisms by which they function.
CD Genomics is committed to combining professional project protocol guidance and standardized analysis processes to ensure that your project is performed accurately and quickly. If you are interested in CD Genomics' CRISPR screening sequencing in the field of genome editing and engineering, please feel free to contact us now.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.

