We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

CRISPR screening technology signifies a transformative advancement in biological research, providing unparalleled capabilities for delineating gene functions, dissecting molecular pathways, and pinpointing therapeutic targets across multiple disciplines. This review examines the extensive applications of CRISPR screening technology within molecular and cellular biology, medical genetics, cancer research, and immunology, highlighting its pivotal role in enhancing both scientific comprehension and clinical interventions.
In recent years, CRISPR screening has seen extensive application in investigating diverse biological processes, encompassing transcriptional regulation, epigenetic mechanisms, protein synthesis, cellular signaling, proliferation, and differentiation. This technology has facilitated the discovery of numerous genes pivotal to these processes.
For instance, Shalem et al. (2014) conducted a comprehensive CRISPR-Cas9 screening study across human cancer cell lines, targeting nearly 18,000 genes. Their research revealed essential genes crucial for cell viability, thus constructing a comprehensive genetic dependency map in cancer cells.
Additionally, CRISPR interference (CRISPRi) has been instrumental in dissecting transcriptional regulatory networks. Horlbeck et al. (2016) employed CRISPRi screens to elucidate the roles of transcription factors (TFs) and cofactors in regulating gene expression in mammalian cells. Their investigation uncovered hierarchical relationships among TFs and identified novel regulatory mechanisms governing cellular differentiation and responses to environmental cues.
CRISPR technology represents a robust tool in medical genetics, presenting significant opportunities for precision medicine. Its application in CRISPR screening allows for the identification of genes linked to human diseases and their genetic variations, particularly ambiguous base mutations within pathogenic genes.
Researchers have employed CRISPR-Cas9 to investigate genetic disorders by inducing targeted mutations in specific genes associated with human ailments. For example, Platt et al. (2014) illustrated the feasibility of CRISPR-Cas9 in rectifying mutations responsible for cystic fibrosis in cells derived from patients. This approach not only corrected the genetic anomaly but also facilitated personalized therapeutic strategies tailored to individual patients.
In the domain of cancer genetics, CRISPR screens have enabled a thorough exploration of genetic susceptibilities across diverse malignancies. Wang et al. (2014) utilized CRISPR-Cas9 screening to pinpoint essential genes crucial for the survival of cancerous cells, thereby uncovering potential targets for therapeutic intervention. Through systematic gene knockouts across various cancer types, the study elucidated the genetic landscapes of tumors and proposed avenues for developing targeted therapeutic approaches.
Take the Next Step: Explore Related Services
CRISPR screening technology is highly pertinent to cancer research, given its utility across a spectrum of phenotypes crucial in oncological investigations. These encompass metastasis, drug resistance, immune evasion, apoptosis inhibition, metabolic reprogramming, genomic instability, and angiogenesis. Moreover, CRISPR screening enables the discovery of synthetic lethal genes, which pose challenges as drug targets. For instance, it elucidates the synthetic lethal effect of PARP1 inhibition in tumors harboring BRCA1/BRCA2 mutations. The technology serves as a versatile tool applicable in diverse research models, including cancer cells derived from various organs, organoids, and animal models.
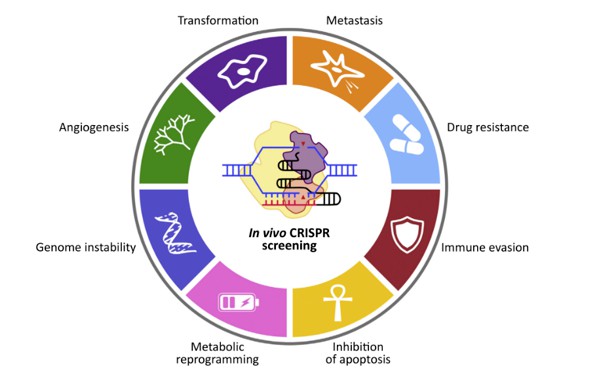 Phenotypes available for CRISPR screening
Phenotypes available for CRISPR screening
CRISPR-Cas9 technology represents a transformative advancement in cancer research, offering a potent mechanism for methodically elucidating the genetic foundations of malignancies. In a seminal investigation, Wang et al. (2014) conducted a comprehensive genome-wide CRISPR screen aimed at pinpointing indispensable genes crucial for the viability of cancerous cells. Through systematic gene knockout experiments across diverse cancer cell lineages, critical vulnerabilities were identified, thereby elucidating novel cancer dependencies of significant therapeutic interest. These discoveries establish a solid framework for the future development of precision-targeted therapeutic strategies.
In a notable study, Shalem et al. (2014) executed an extensive CRISPR-Cas9 screen aimed at identifying genes influencing the responsiveness of melanoma cells to vemurafenib, a BRAF inhibitor. This investigation authenticated established mechanisms of resistance while revealing novel genetic determinants implicated in drug resistance. Through elucidation of intricate genetic frameworks underpinning therapeutic resistance, the inquiry provided fresh perspectives on prospective combinatorial therapies for mitigating resistance in melanoma management.
Parnas et al. (2015) illustrated the efficacy of CRISPR screens in discerning genes implicated in the immune response against cancer. Through targeted gene knockout in dendritic cells using CRISPR-Cas9, pivotal regulators of the tumor necrosis factor (TNF) reaction to lipopolysaccharide (LPS) were identified. Emphasis was placed on the involvement of the PAF and OST complexes in modulating immune responses, proposing novel strategies for augmenting anti-tumor immunity via genetic interventions.
CRISPR-Cas9 technology has markedly progressed immunological research, particularly in elucidating intricate regulatory networks governing immune responses. A seminal investigation by Parnas et al. (2015) exemplified the potency of genome-wide CRISPR screens in primary immune cells, specifically dendritic cells (DCs). Through the deployment of a comprehensive pooled CRISPR-Cas9 library into DCs, coupled with stimulation by bacterial lipopolysaccharide (LPS), numerous genes governing the induction of tumor necrosis factor (TNF), pivotal in immune response to pathogens, were delineated. This methodology facilitated the identification of established TLR4 signaling regulators alongside novel gene candidates, thereby enhancing comprehension of innate immune mechanisms.
Parnas et al. (2015) categorized the identified genes into three functional modules with distinct effects on lipopolysaccharide (LPS)-induced tumor necrosis factor (TNF) production. The study highlighted the involvement of the PAF and OST complexes, previously unrelated to TLR4 signaling. Disruption of genes within these complexes revealed substantial impacts on TNF response, indicating novel regulatory dimensions within the TLR4 pathway. This discovery underscores the efficacy of CRISPR screening in revealing unforeseen elements of established pathways, thereby enhancing comprehension of immune regulation.
CRISPR screening in immunology plays a pivotal role in identifying potential targets for immunotherapy. For example, Shalem et al. (2014) performed genome-wide CRISPR screens to investigate the modulation of immune checkpoint pathways in cancer treatment. Through gene knockout experiments in T cells and subsequent analysis of their interactions with tumor cells, the study unveiled essential regulators of T cell activation and exhaustion. These findings hold significant promise for optimizing immunotherapeutic approaches, particularly checkpoint inhibitors, through targeted manipulation of distinct genetic pathways.
CRISPR screens have played a pivotal role in unraveling the genetic underpinnings of autoimmune diseases. In a study by Wang et al. (2014), a genome-wide CRISPR screen was conducted to ascertain genes influencing the inflammatory response in macrophages. This investigation identified several novel regulators of cytokine production, offering promising targets for therapeutic interventions in autoimmune disorders. Through systematic gene disruptions and subsequent analysis of cytokine profiles, critical pathways implicated in autoimmune pathogenesis can be pinpointed, paving the way for more targeted treatment strategies.
In the biomedical realm, CRISPR screening technology holds immense potential. Its integration with complementary sequencing techniques, such as single-cell sequencing, enhances research depth and rigor, elevating scholarly standards across disciplines. At CD Genomics, we leverage CRISPR screens to elucidate intricate genetic regulations, identify innovative therapeutic targets, and advance precision medicine initiatives. As CRISPR technology continues to evolve, its transformative impact on biomedical research and clinical applications remains unparalleled.
References

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy