What is Transcription Factor Analysis?
Transcription factor research is one of the important components of molecular regulatory research, which itself is a class of proteins that can bind directly to DNA sequences and specifically regulate gene expression upstream of the molecular machinery. And in a biological sense, transcription factors play an important regulatory role in the growth and development of living organisms and their response to the outside world. Among them, analyzing the types, functional roles, influence strengths and binding sites (TFBS) of transcription factors is the key to transcription factor research. In the study of transcription factors based on TFBS, ChIP-seq is usually utilized to mine TFBS and related target genes, but due to the limitation of antibodies, ChIP-seq cannot work well in non-model organisms. At this time, another assay becomes the first choice for TFBS research in non-modal organisms, which is DAP-seq. In actual research, a transcriptome assay is usually added along with DAP-seq to form a transcriptome + DAP-seq multi-omics association scheme.
What is DAP-seq?
DAP-seq can be regarded as a kind of expansion and depth of "transcription factor analysis" in transcriptome analysis; DAP-seq studies the DNA binding sequence and potential target genes of a certain transcription factor, while RNA-seq studies the differential genes among samples. DAP-seq studies the DNA binding sequences and potential target genes of a transcription factor, while RNA-seq studies the differential genes among samples. In addition, DAP-seq is a specific experiment-based genomics tool that identifies regulatory target genes more accurately. Transcriptomics and DAP-seq complement each other in studying the transcription factor-target gene regulatory network relationship, and can accurately dig out the core genes with significant contribution to the phenotype.
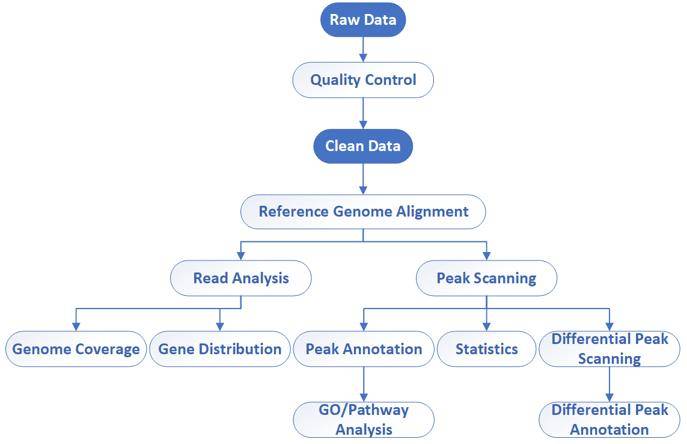 Bioinformatics workflow of DAP-Seq – CD Genomics
Bioinformatics workflow of DAP-Seq – CD Genomics
Please refer to our service DAP-Seq Service (DNA affinity purification sequencing).
If you want to know more about DAP-seq, please read our article What is DAP-Seq.
Case Study: Regulatory Mechanism of Transcription Factor Cswrky33 for Disease Resistance to Citrus Green Mold
The authors conducted a transcriptome assay to identify transcription factors that respond significantly to citrus infection with green mold, and finally identified CsWRKY33 as a key transcription factor in the disease resistance mechanism of citrus.
Then, in order to identify the DNA binding site of CsWRKY33 in the genome, and then to identify the corresponding target genes, the authors used the DAP-seq technique to identify the target genes related to CsWRKY33. The DAP-seq technique was used to identify the target genes related to CsWRKY33, i.e., the CsWRKY33 transcription factor was expressed in vitro, and the sequence recognition-binding-precipitation reaction was carried out with the genome extracted from citrus as the reaction substrate. The precipitated sequences are then up-converted and the relevant target genes that bind to CsWRKY33 are thus identified.
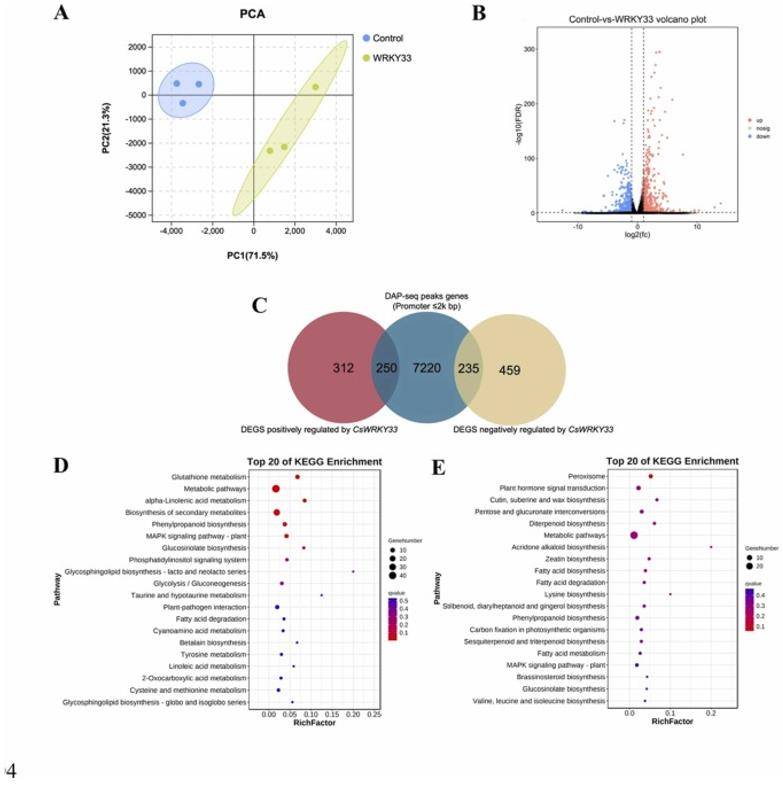 Transcriptomics and DAP-seq analysis. (Wang et al., 2023)
Transcriptomics and DAP-seq analysis. (Wang et al., 2023)
In DAP-seq, the sequence information, type and distribution of CsWRKY33-related binding sites, as well as the analysis of significantly enriched motif sequences, can be obtained. From the above results, it was found that CsWRKY33 is a typical transcription factor that binds to DNA promoters and regulates gene activity, and its main binding site is W-box (a highly conserved region). And a total of 40,365 peak genes were identified with an average length of 294 bp, accounting for 3.63% of the genome.
Reference:
- Wang, Wenjun, et al. "A self-regulated transcription factor CsWRKY33 enhances resistance of citrus fruit to Penicillium digitatum." Postharvest Biology and Technology 198 (2023): 112267.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Bioinformatics workflow of DAP-Seq – CD Genomics
Bioinformatics workflow of DAP-Seq – CD Genomics Transcriptomics and DAP-seq analysis. (Wang et al., 2023)
Transcriptomics and DAP-seq analysis. (Wang et al., 2023)