CRISPR technology has significantly transformed genetic engineering, providing unprecedented capabilities for precise genome editing. However, the potential for off-target effects poses critical challenges that require thorough investigation and validation. This article delves into CRISPR off-target analysis, elucidating the implications, scoring systems, prediction methods, and validation techniques involved. CD Genomics remains at the forefront of these analyses, offering advanced solutions to mitigate risks associated with off-target effects.
What is the Off-Target Effect?
The off-target effect in CRISPR refers to unintended edits in the genome that occur when the CRISPR-Cas9 system interacts with DNA sequences other than the intended target. This phenomenon arises when the guide RNA (gRNA) recognizes and binds to sequences that are similar but not identical to the target site. Research indicates that the specificity of the CRISPR system is influenced by the length of the gRNA and the presence of a protospacer adjacent motif (PAM) sequence, both of which play crucial roles in determining binding affinity.
Studies have demonstrated that off-target effects can lead to mutations in critical genomic regions, potentially resulting in adverse biological consequences. For instance, a study published in Nature highlighted that Cas9 could induce mutations at off-target sites that share as little as three nucleotides with the intended target. Such mutations could disrupt essential genes, leading to oncogenesis or other disease states, emphasizing the need for rigorous off-target analysis.
What Happens If CRISPR is Off-Target?
When CRISPR technology induces off-target effects, the implications can be profound and far-reaching, impacting both research outcomes and potential therapeutic applications. Understanding the various consequences of off-target mutations is critical for assessing the overall safety and efficacy of CRISPR-based interventions.
1. Disruption of Essential Genes
One of the most significant risks associated with off-target effects is the inadvertent alteration of essential genes. When CRISPR-Cas9 makes unintended edits, it may disrupt the function of genes critical for normal cellular processes. For instance, mutations in tumor suppressor genes like TP53 or PTEN can lead to uncontrolled cell proliferation, thereby increasing the risk of cancer. A study published in Nature Biotechnology revealed that CRISPR-induced mutations in TP53 resulted in loss of function, contributing to oncogenic pathways in several models.
2. Unintended Phenotypic Changes
Off-target mutations can lead to unintended phenotypic alterations that may not be immediately apparent. Such changes can complicate the interpretation of experimental results, making it difficult to determine whether observed effects are due to intended CRISPR edits or off-target events. For example, a study in Cell highlighted that even subtle off-target modifications in regulatory regions could lead to significant changes in gene expression, ultimately affecting cellular behavior and phenotype.
3. Implications for Therapeutic Applications
In the context of gene therapy, off-target effects can pose severe safety risks. For instance, if CRISPR is used to treat genetic disorders by correcting mutations, unintended edits elsewhere in the genome could introduce new health issues. Research has indicated that off-target effects could lead to the activation of oncogenes or the silencing of essential genes, complicating treatment outcomes. For example, if CRISPR is employed to correct a mutation in a hematopoietic stem cell, unintended modifications could affect the cell's ability to differentiate properly, resulting in hematological disorders.
4. Ethical and Regulatory Concerns
The potential for off-target effects raises significant ethical and regulatory questions regarding the use of CRISPR in clinical settings. The unpredictability associated with off-target mutations can lead to public apprehension about the safety of genetic editing technologies. Regulatory bodies, such as the U.S. Food and Drug Administration (FDA), require comprehensive evaluations of off-target effects before allowing CRISPR-based therapies to proceed to clinical trials. This necessitates robust off-target analysis protocols that can reliably identify and quantify unintended edits.
5. Impact on Research Validity
Off-target effects can severely impact the validity of scientific research employing CRISPR technology. If off-target mutations go undetected, they may confound results, leading to erroneous conclusions about the role of specific genes or pathways. This could undermine the reproducibility of studies and create misleading data that can propagate through the scientific literature. A notable instance involved studies where CRISPR was used to interrogate gene function, only to discover later that observed phenotypes were attributed to off-target effects rather than the intended gene modifications.
6. Long-term Consequences
Long-term consequences of off-target effects remain an area of active research. Unintended edits may not produce immediate observable effects but can lead to cumulative changes over time. Such delayed consequences could manifest as late-onset diseases or altered cellular behaviors in subsequent generations of cells. This is particularly concerning in applications involving germline editing, where changes are heritable. Studies have called for rigorous long-term studies to understand how off-target effects could impact not only individual subjects but also future generations.
Off-Target Score: A Measure of Potential Risk
The off-target score is a quantitative measure used to evaluate the likelihood of unintended targeting by the CRISPR-Cas9 system. This score is derived from several factors, including:
1. Sequence Homology: The degree of similarity between the gRNA and potential off-target sites is assessed.
2. PAM Sequence Compatibility: The presence of a suitable PAM sequence adjacent to the target site is critical for Cas9 binding.
3. Local Sequence Context: The genomic environment surrounding the potential off-target site can influence binding affinity and specificity.
A higher off-target score suggests an increased risk of unintended edits, making it essential for researchers to consider these scores during gRNA design. CD Genomics utilizes advanced bioinformatics tools to calculate off-target scores, ensuring that researchers can make informed decisions regarding gRNA selection.
Methods for Off-Target Analysis in CRISPR
Off-target analysis encompasses various methodologies employed to detect and quantify unintended edits post-CRISPR editing. These methods include:
1. In Silico Prediction
Bioinformatics tools facilitate the identification of potential off-target sites by analyzing the gRNA sequence against the entire genome. Tools such as Cas-OFFinder and CRISPRseek employ algorithms to predict off-target sites based on sequence homology and PAM compatibility. These predictions allow researchers to prioritize potential off-targets for further experimental validation.
2. High-Throughput Sequencing Techniques
Next-generation sequencing (NGS) technologies are instrumental in identifying off-target effects. Techniques such as Digenome-seq provides comprehensive profiling of off-target mutations:
Digenome-seq: In this approach, genomic DNA is treated with CRISPR-Cas9, and the resulting cleavage products are analyzed to reveal off-target modifications based on the cleavage pattern.
These high-throughput methods offer a detailed overview of off-target effects, enhancing the reliability of CRISPR applications.
3. PCR and Sanger Sequencing
For specific off-target sites identified through predictive models, polymerase chain reaction (PCR) amplification followed by Sanger sequencing can confirm the presence of unintended edits. This targeted approach enables researchers to ascertain the frequency and nature of off-target mutations.
You may interested in
Learn More
Strategies to Reduce Off-Target Effects
Reducing off-target effects is critical for improving the safety and efficacy of CRISPR-based gene editing technologies. Various strategies can be employed to minimize unintended mutations, ranging from optimizing guide RNA design to implementing advanced delivery methods. This section outlines key strategies to mitigate off-target effects effectively.
1. Design Optimization of Guide RNAs
The design of gRNAs is one of the most crucial factors influencing off-target activity. The following strategies can enhance gRNA specificity:
a. Target Sequence Selection
Choosing target sequences that are unique to the gene of interest is fundamental. Researchers can employ bioinformatics tools to analyze potential off-target sites across the genome. Tools like CRISPOR and CCTop provide valuable insights by predicting off-target effects based on sequence complementarity and mismatches, allowing for more informed gRNA design.
b. Truncated gRNAs
Using truncated gRNAs, which contain shorter lengths of complementary sequences, can help improve specificity. A study published in Nature Communications demonstrated that 17- to 18-nucleotide-long gRNAs exhibited reduced off-target effects compared to standard 20-nucleotide gRNAs, while still maintaining sufficient on-target activity.
2. Enhanced Cas Protein Variants
The choice of Cas protein can significantly influence the precision of CRISPR edits. Several engineered variants of Cas9 exhibit enhanced specificity:
a. High-Fidelity Cas9 Variants
Variants like SpCas9-HF1 and eSpCas9 have been developed to reduce off-target effects. These variants are engineered to have reduced non-specific binding without compromising on-target efficiency. For instance, research has shown that eSpCas9 significantly lowers off-target activity while retaining effective gene editing capabilities.
b. Cas12 and Cas13 Systems
Alternative CRISPR systems, such as Cas12 and Cas13, offer different target recognition mechanisms that can inherently reduce off-target effects. Cas12, for instance, has been shown to exhibit a lower rate of off-target cleavage due to its unique recognition of target DNA, which enhances specificity compared to traditional Cas9.
3. Conditional and Controlled Delivery Methods
The delivery method of CRISPR components can also impact off-target effects. Using refined delivery techniques can enhance the precision of gene editing:
a. Lentiviral Vectors
Lentiviral vectors can be employed to deliver CRISPR components in a controlled manner, allowing for stable integration of genetic modifications. This method can help reduce off-target effects by ensuring that the CRISPR machinery is expressed at optimal levels and durations, minimizing the risk of off-target cleavage.
b. Nanoparticle-Based Delivery
Nanoparticle-based delivery systems provide a promising avenue for targeted delivery of CRISPR components. These systems can be engineered to facilitate cellular uptake while minimizing exposure to non-target tissues, thus reducing the likelihood of off-target effects. Studies have shown that lipid nanoparticles can effectively deliver CRISPR components with minimal off-target cleavage.
4. Screening and Validation Techniques
Employing robust screening and validation techniques can help identify and mitigate off-target effects post-editing:
a. Deep Sequencing Technologies
NGS techniques can be used to analyze off-target effects comprehensively. By sequencing potential off-target sites, researchers can identify unintended mutations and quantify their frequency. For instance, a study in Nature Biotechnology utilized NGS to validate off-target effects and demonstrated the ability to characterize CRISPR-induced mutations across the genome.
b. In Vivo Models
Using in vivo models can provide critical insights into the biological relevance of off-target effects. By assessing the phenotypic and functional consequences of CRISPR editing in live organisms, researchers can better understand the implications of off-target mutations. This approach is particularly important in the context of therapeutic applications where potential risks must be thoroughly evaluated.
5. Post-Editing Analysis and Repair Mechanisms
Even with careful design and delivery, some off-target effects may still occur. Implementing repair mechanisms can help rectify unintended edits:
a. Base Editing Technologies
Base editing allows for precise nucleotide changes without introducing double-strand breaks, minimizing the likelihood of off-target effects. This method utilizes a modified Cas9 fused to a deaminase enzyme, enabling direct conversion of one DNA base into another. Studies have indicated that base editing produces fewer off-target mutations compared to traditional CRISPR-Cas9 approaches, offering a refined strategy for gene modification.
b. Transcription Activator-Like Effector Nucleases (TALENs)
TALENs are another genome editing technology that can be utilized to complement CRISPR systems. These nucleases allow for highly specific DNA cleavage, and when combined with CRISPR, they can provide a dual strategy to counteract potential off-target effects. The unique binding domains of TALENs can be engineered to target specific sequences, offering an additional layer of precision.
Predicting CRISPR Off-Target Effects
Predicting off-target effects is a critical aspect of CRISPR technology that significantly influences the safety and efficacy of gene editing applications. Accurate predictions can guide researchers in the design of more specific gRNAs and reduce the likelihood of unintended mutations in the genome. This section delves into the methodologies and tools used for predicting off-target effects in CRISPR systems.
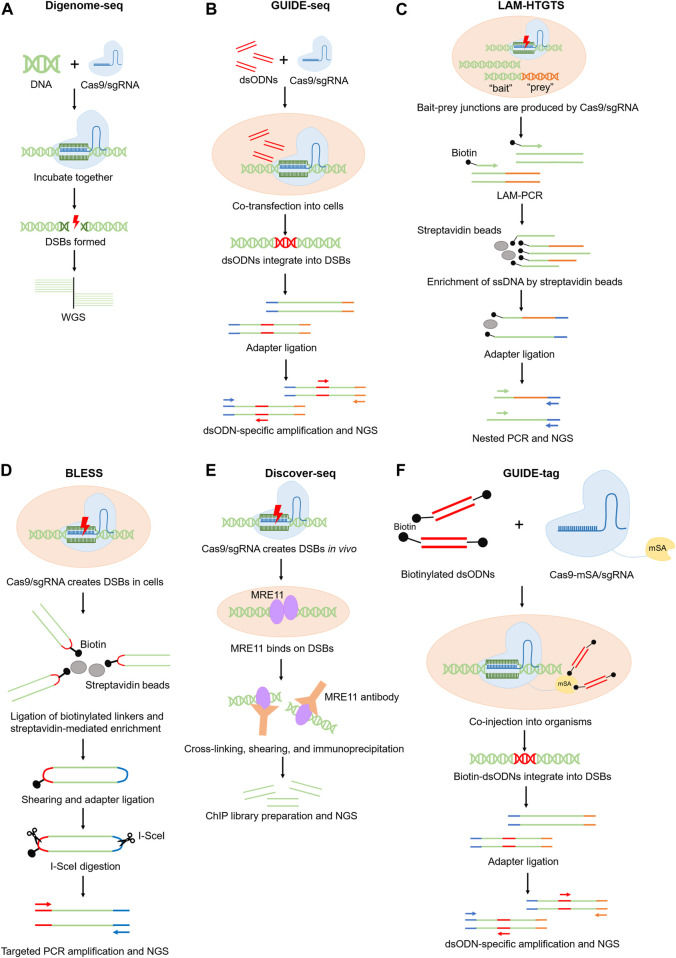 FIGURE 1 Schematics of experimental methods for genome-wide off-target prediction.
FIGURE 1 Schematics of experimental methods for genome-wide off-target prediction.
Computational Tools for Off-Target Prediction
Several computational tools have been developed to predict off-target effects, utilizing various algorithms that assess the likelihood of gRNA binding to non-target sites in the genome. Here are some prominent tools in this field:
1. CRISPOR
CRISPOR is a widely used web-based tool that allows researchers to design and evaluate gRNAs. It predicts potential off-target sites based on sequence complementarity and mismatch tolerance. CRISPOR calculates an off-target score that reflects the likelihood of binding, taking into account various factors such as:
- Number of mismatches: The greater the number of mismatches between the gRNA and potential off-target sequences, the lower the likelihood of off-target activity.
- PAM sequence compatibility: The presence of the protospacer adjacent motif (PAM) is essential for Cas9 binding, and CRISPOR evaluates PAM compatibility to enhance prediction accuracy.
2. CCTop
CCTop (CRISPR/Cas9 target online predictor) is another effective tool that provides predictions for potential off-target sites. CCTop allows users to input target sequences and generates a list of potential off-target sites within the genome. The platform evaluates the gRNA's binding efficiency by considering:
- Mismatches and bulges: The tool assesses various types of mismatches (G/C to A/T) and bulges, which can affect binding specificity.
- Genomic context: CCTop incorporates the local genomic context to provide a more accurate prediction of off-target interactions.
Machine Learning Approaches
Machine learning has emerged as a powerful approach for predicting CRISPR off-target effects by leveraging large datasets of known CRISPR interactions:
a. Deep Learning Models
Recent advancements in deep learning have led to the development of models that predict off-target effects based on sequence data. These models utilize neural networks trained on extensive datasets, capturing complex relationships between gRNA sequences and their off-target potential. For example, the DeepCRISPR framework employs a deep learning architecture to predict both on-target and off-target cleavage sites with high accuracy.
b. Random Forest Algorithms
Random forest algorithms are also used to create predictive models for off-target effects. By analyzing various sequence features, including mismatches, bulges, and local genomic context, these models can classify the likelihood of off-target activity. Such algorithms have shown promising results in providing reliable predictions, facilitating the selection of high-specificity gRNAs.
Experimental Validation of Predictions
While computational predictions are valuable, experimental validation remains crucial for confirming off-target effects. Various techniques can be employed to validate predictions and assess the actual outcomes of CRISPR editing.
Integration of Prediction and Validation
The integration of computational predictions with experimental validation provides a robust framework for assessing and mitigating off-target effects in CRISPR applications. Researchers can utilize predictive tools to design gRNAs with minimal off-target potential and subsequently confirm these predictions through experimental techniques. This synergistic approach is critical for enhancing the safety and efficacy of CRISPR-based therapies, particularly in clinical settings.
Predicting CRISPR off-target effects is a multifaceted process that combines computational modeling, machine learning, and experimental validation. The use of advanced tools and algorithms, such as CRISPOR and CCTop, enables researchers to identify potential off-target sites efficiently. As CRISPR technology continues to evolve, refining prediction methodologies will play a pivotal role in advancing the precision and safety of gene editing applications, aligning with CD Genomics' commitment to excellence in genomic research and development.
Validating CRISPR Off-Target Effects
Validating CRISPR off-target effects is a critical step in ensuring the safety and efficacy of gene editing applications. The process involves confirming the unintended modifications predicted or detected through initial screening, using various experimental techniques to provide robust evidence of off-target interactions. This section discusses several key methodologies employed for the validation of CRISPR off-target effects, including conventional approaches like deep sequencing, as well as more specialized techniques such as multiplex PCR and hybrid capture.
Importance of Validation
Validation of off-target effects is crucial for several reasons:
- Safety Assessment: Unintended genetic modifications can lead to potential health risks, especially in therapeutic applications. Validating off-target effects helps assess the safety profile of CRISPR interventions.
- Optimization of gRNA Design: Validation can provide insights into the design of gRNAs that minimize off-target activity, enhancing the precision of gene editing.
- Regulatory Compliance: For clinical applications, validation of off-target effects is often required to meet regulatory standards and ensure responsible use of gene editing technologies.
Methodologies for Validating Off-Target Effects
Several methodologies are employed to validate off-target effects detected in CRISPR systems. These include:
1. Deep Sequencing
Deep sequencing remains one of the most powerful techniques for validating off-target effects. This high-throughput method allows for comprehensive analysis of the genome and can detect low-frequency mutations. The validation process typically involves:
1. Targeting with CRISPR: The CRISPR-Cas9 system is used to target specific genomic sites.
2. PCR Amplification: Amplification of both target and potential off-target regions is performed.
3. Sequencing: The amplified products are sequenced to identify any modifications, including those at predicted off-target sites.
Deep sequencing provides a quantitative measure of off-target activity, allowing researchers to determine the frequency and nature of off-target mutations.
2. Multiplex PCR
Multiplex PCR is an effective validation technique that enables the simultaneous amplification of multiple target sequences, including both on-target and off-target sites. This technique involves:
1. Designing Specific Primers: Primers specific to both on-target and potential off-target sites are designed.
2. PCR Amplification: The multiplex PCR reaction amplifies all targeted regions in a single reaction, allowing for efficient analysis.
3. Gel Electrophoresis or Sequencing: The amplified products are analyzed through gel electrophoresis or next-generation sequencing to detect mutations at off-target sites.
Multiplex PCR offers a cost-effective and time-efficient approach to validate multiple off-target effects simultaneously, making it an attractive option for high-throughput studies.
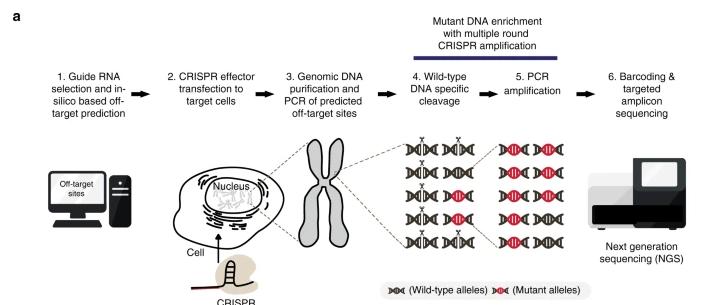 FIGURE 2 Schematics of the detection of off-target mutations using the CRISPR amplification method.
FIGURE 2 Schematics of the detection of off-target mutations using the CRISPR amplification method.
3. Hybrid Capture
Hybrid capture is a sophisticated technique that combines the specificity of hybridization with high-throughput sequencing to validate off-target effects. The steps involved in this process include:
1. Designing Capture Probes: Specific probes are designed to hybridize with both on-target and off-target sequences across the genome.
2. Hybridization: The genomic DNA is incubated with the capture probes, allowing for the formation of DNA-probe complexes.
3. Isolation and Sequencing: The hybridized complexes are captured, followed by sequencing to identify any off-target modifications.
Hybrid capture allows for targeted enrichment of regions of interest, facilitating the validation of off-target effects with high sensitivity and specificity.
Factors Influencing Validation
Several factors can influence the success of off-target validation, including:
- gRNA Design: The design of the gRNA can significantly impact off-target effects, influencing the choice of validation methods and their sensitivity.
- Cell Type and Context: The cellular context may affect the expression levels of CRISPR components and the likelihood of off-target interactions, necessitating validation in relevant cell types.
- Depth of Sequencing: The depth of sequencing in methods like deep sequencing can determine the ability to detect low-frequency off-target events.
Validating CRISPR off-target effects is an essential component of ensuring the safety and effectiveness of gene editing technologies. By employing a combination of methodologies such as deep sequencing, multiplex PCR, and hybrid capture, researchers can comprehensively assess off-target interactions and their implications. This rigorous validation process aligns with CD Genomics' commitment to advancing genomic research responsibly, paving the way for more precise and effective applications of CRISPR technology in therapeutic and research contexts.
Summary
The analysis of CRISPR off-target effects is integral to advancing the safety and efficacy of genome editing technologies. Understanding the mechanisms underlying off-target events, employing robust predictive and analytical methodologies, and implementing effective mitigation strategies are essential for the responsible application of CRISPR in research and therapeutic settings. CD Genomics remains dedicated to providing comprehensive solutions for off-target analysis, ensuring that CRISPR technologies can be harnessed effectively and safely in the future.
References:
- Guo C, Ma X, Gao F, Guo Y. Off-target effects in CRISPR/Cas9 gene editing. Front Bioeng Biotechnol. 2023
- Chen, Q., Chuai, G., Zhang, H. et al. Genome-wide CRISPR off-target prediction and optimization using RNA-DNA interaction fingerprints. Nat Commun 14, 7521 (2023).
- Kang, SH., Lee, Wj., An, JH. et al. Prediction-based highly sensitive CRISPR off-target validation using target-specific DNA enrichment. Nat Commun 11, 3596 (2020).


 Sample Submission Guidelines
Sample Submission Guidelines
 FIGURE 1 Schematics of experimental methods for genome-wide off-target prediction.
FIGURE 1 Schematics of experimental methods for genome-wide off-target prediction. FIGURE 2 Schematics of the detection of off-target mutations using the CRISPR amplification method.
FIGURE 2 Schematics of the detection of off-target mutations using the CRISPR amplification method.