Genome-wide association analysis (GWAS) serves as a potent tool for pinpointing genes associated with traits, playing a pivotal role in identifying candidate genomic loci influencing phenotypic variations. However, conventional GWAS primarily relies on information from single nucleotide variants (SNPs) to establish associations with phenotypes, neglecting substantial structural variants (SVs) with considerably larger phenotypic effect sizes. Although the overall number of SVs in the genome is fewer than that of SNPs, SVs encompass a more extensive span of bases and influence a significantly larger total number of genomic sequence variations than SNPs.
In contrast to sequence polymorphic variants, structural variants may exert a more profound impact on the genomes and genetic traits of plants and animals. Notably, SV-based GWAS analyses have found widespread application in various species, including maize, cucumber, grape, wheat, and tomato.
Genome of Tomato
In a recent publication in Nature Genetics titled "Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species," they compiled 11 chromosome-level, high-quality genomes from wild and cultivated tomato varieties. This effort led to the creation of the world's first super-pangenome for tomatoes, facilitating a groundbreaking SV-GWAS analysis based on the pangenome profiles. The outcomes of this study are poised to significantly expedite biological research and the breeding of tomatoes, a globally important crop.
A total of 11 tomato varieties, including 8 wild-type variants, 1 near-wild tomato species, and 2 cultivated varieties, were procured for this study. Employing advanced sequencing technologies such as PacBio and Hi-C, the genomes of these 11 tomato varieties were meticulously assembled at the chromosome level. Subsequently, a super-pan-genome was constructed, integrating the genetic profiles of all 11 tomato genomes. This consolidated genome served as the basis for analyzing structural variations in the context of structural variants (SV).
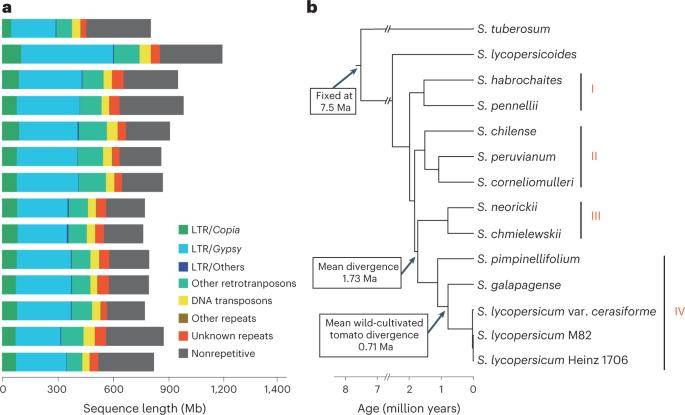 Phylogenetic relationships and genomic components of wild and domesticated tomatoes. (Li et al., 2023)
Phylogenetic relationships and genomic components of wild and domesticated tomatoes. (Li et al., 2023)
Super-pangenome of Tomatoes
The investigation pioneered the assembly of a comprehensive super pangenome for tomatoes, encompassing 11 Solanum species. Leveraging cluster analysis, the study unveiled 40,457 pan gene families, aggregating protein-coding genes from 11 chromosome-level genomes and two previously published ones. From the reported tomato pan-genome, 3,441 of the 4,874 non-intronic genes were incorporated, while an additional 9,320 nonredundant genes absent in the reported pan-genome were identified.
Comparative genome analysis exposed 2.0-8.193 billion SNPs and 0.6-1.5 million small InDels (≤50 bp) across 12 tomato genomes. Furthermore, the study identified 103,333 insertions, 119,794 deletions, 41,960 CNVs, 23,516 translocations, and 1,320 retrogrades (< 1 Mb in length) within the 12 tomato germplasms. Notably, the study accurately detected previously reported SVs associated with phenotypic variation. In a departure from previous tomato pan-SV mapping, the study exclusively identified 180,314 SVs, contributing to a total of 224,447 SVs.
Integration of these identified SVs with a pan-SV dataset (comprising 112 tomato germplasms) facilitated the exploration of SV differences in tomato evolution. Subdividing the 112 cultivars into four groups revealed that the majority of SVs occurred at low frequencies (less than 0.25) across all groups. Intriguingly, 8,094 SVs exhibited significant frequency variations between wild and cultivated groups. Functional analysis highlighted the involvement of these genes in processes such as meristematic tissue development and ammonium transport. Additionally, 388 highly differentiated SVs were pinpointed between wild and cultivated tomatoes, adding a nuanced layer to our understanding of tomato genetic evolution.
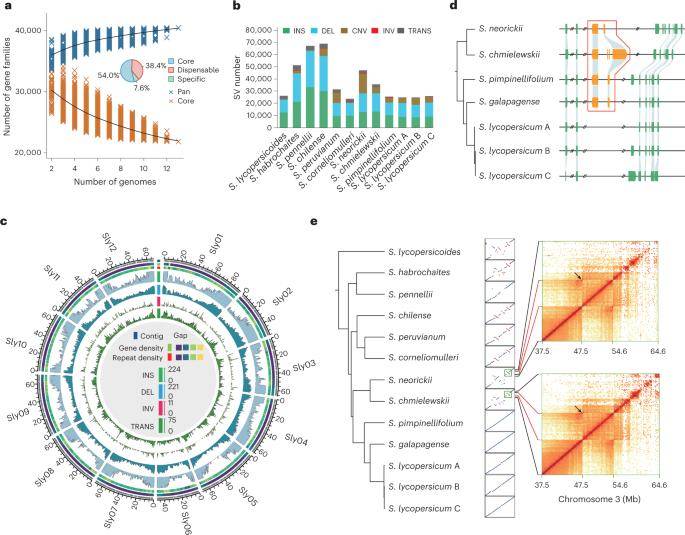 Super-pangenome and the landscape of structural variation among wild and cultivated tomatoes. (Li et al., 2023)
Super-pangenome and the landscape of structural variation among wild and cultivated tomatoes. (Li et al., 2023)
Uncovering Crucial Genes Enhancing Tomato Yield
In the comparison between wild and cultivated tomato varieties, this investigation pinpointed 388 structural variations (SVs) that significantly influenced 278 genes. Notably, a noteworthy alteration in the frequency of a 244-base pair deletion took the second position, manifesting in the initial exon of the Sgal12g015720 gene. This gene encodes a protein belonging to the cytochrome P450 (CYP) superfamily, known for its pivotal role in plant growth, development, and the biosynthesis of secondary metabolites. Sgal12g015720 exhibited peak expression levels in the stems of the wild tomato, S. pennellii, whereas its expression was nearly negligible in the two cultivated tomato species.
The occurrence of the 244-base pair deletion event is hypothesized to be a byproduct of tomato domestication, resulting in the pseudogenization of the Sgal12g015720 gene in cultivated tomatoes. This event holds potential significance in the regulation of plant size and yield during the course of tomato breeding. Subsequent studies corroborated this hypothesis through experimental analyses involving the overexpression of the gene. The findings underscore the pivotal role of Sgal12g015720 in shaping tomato yield, shedding light on its potential application in future breeding strategies.
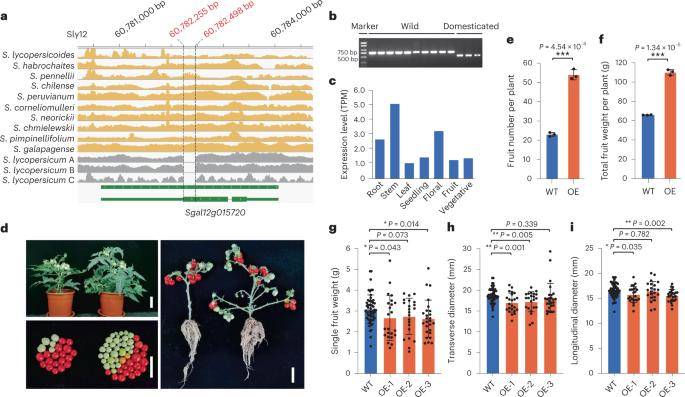 Characterization of a wild tomato cytochrome P450 gene, Sgal12g015720. (Li et al., 2023)
Characterization of a wild tomato cytochrome P450 gene, Sgal12g015720. (Li et al., 2023)
This study presents the chromosome-level genomes of nine wild tomato species and two cultivars, amalgamating them with two previously published genomes to unveil the phylogeny of the tomato. Through this comprehensive approach, a super-pan-genome for tomatoes was constructed. The investigation delved into the structural variations within the tomato genome, shedding light on the genomic diversity inherent in the wild relatives of tomatoes. Notably, the study led to the identification of a wild tomato gene with promising potential to enhance yield in contemporary cultivated tomatoes.
The map-based genome construction served as a foundation for conducting structural variation (SV)-based genome-wide association studies, revealing numerous signals intricately linked to flavor-related traits and fruit metabolites in tomatoes. This holistic exploration provides valuable insights into the genetic factors influencing tomato characteristics, offering a pathway for leveraging this knowledge in the pursuit of improved crop yields and enhanced flavor profiles.
Reference:
- Li, N., He, Q., Wang, J. et al. Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species. Nat Genet 55, 852–860 (2023).
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Phylogenetic relationships and genomic components of wild and domesticated tomatoes. (Li et al., 2023)
Phylogenetic relationships and genomic components of wild and domesticated tomatoes. (Li et al., 2023) Super-pangenome and the landscape of structural variation among wild and cultivated tomatoes. (Li et al., 2023)
Super-pangenome and the landscape of structural variation among wild and cultivated tomatoes. (Li et al., 2023) Characterization of a wild tomato cytochrome P450 gene, Sgal12g015720. (Li et al., 2023)
Characterization of a wild tomato cytochrome P450 gene, Sgal12g015720. (Li et al., 2023)