Urine Microbiome Research Solution
Inquiry
>
The Importance of Detecting Urine Microbiome
Compared to many other microbial niches, the human urinary tract harbors relatively low biomass. Studies have identified many genera and species that may constitute a core urinary microbiome, which mainly varies between male and female, the young and the old, patients and the healthy. Typically, the biomass of the human urine microbiome ranges from 100 to 105 colony-forming units per milliliter of urine. Recent studies have shown that bacteria belonging to the Corynebacterium group are more likely to be found in men; while women are more likely to have bacteria of the genus Lactobacillus that are also found in the human gut. Therefore, understanding the characteristics of urinary microbiome and ecology is critical for diagnostic, prognostic, and therapeutic research, and help to measure, restore, and/or preserve the native, healthy ecology of the urinary microbiomes.
The ability to measure and characterize the urinary microbiome has been enabled through the development of next-generation sequencing and bioinformatic platforms that allow for the unbiased detection of resident microbial DNA.
 Request a Quote
Request a Quote
Accelerate Research and Practice in Urine Microbiome
Based on our next-generation sequencing (NGS), PacBio SMRT sequencing, and Nanopore sequencing platforms, we have tested and analyzed various urinary microbiome samples. We can help you identify the microorganisms presented in the urinary tract, their relative abundance, and evolutionary relationships. We also study the correlation between urinary microbes and human health, the function of urinary microorganisms from gene level can also be studied, all of which are of great importance for scientific research, the development of supplements and therapies.
The Main Research Direction of Urine Microbiome
Exploring the differences in microbial composition and content between the disease group and the healthy group to search for disease-related markers
Studying and comparing the differences of urinary flora structure between individuals at different intervention stages
Studying changes in urinary flora of same individual at different developmental stages
Studying the changes in urinary flora during the development of a certain disease, so as to search for biomarkers
What Can We Do?
-
1. Identification and discovery of urinary microorganisms
-
2. Quantitation of microorganisms in the urinary tract
-
3. Evolutionary and functional analysis of microorganisms in the urinary tract
-
4. Relationship between microorganisms and diseases
Note: Our services are for research use only, not for disease diagnosis and treatment.
Detectable Objects
We have successfully analyzed reproductive urine microbiome samples from human and other animals.
Detectable Microorganisms
Fungi, bacteria, and viruses, etc.
Technical Platforms
We are equipped with Illumina HiSeq/MiSeq, Ion PGM, PacBio SMRT systems, Nanopore systems, PCR-DGGE (PCR-denaturing gradient gel electrophoresis), real-time qPCR, clone library, and other detection platforms.
Sample Requirement
-

- DNA sample: DNA ≥ 500ng, concentration ≥ 10ng/ul, and OD260/280 = 1.8-2.0.
-
Ensure the DNA is not degraded or slightly degraded and avoid repeated freezing and thawing cycles.
-
Please use enough ice packs or dry ice during transportation.
Workflow
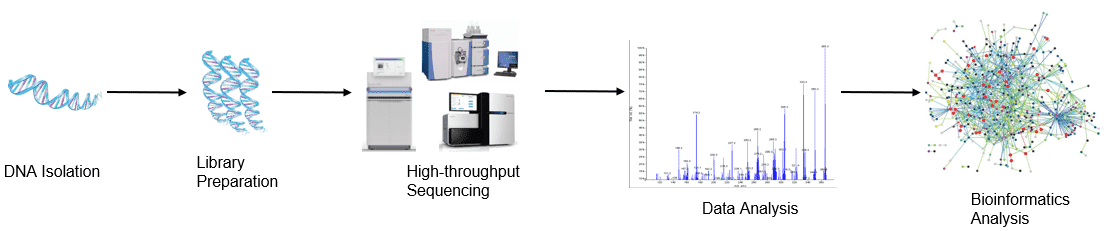 Figure 1. High-throughput sequencing analysis process
Figure 1. High-throughput sequencing analysis process
 Figure 2. PCR-DGGE analysis process
Figure 2. PCR-DGGE analysis process
Bioinformatics Analysis
Our bioinformatics analyses are flexible to your needs.
|
OTU Clustering |
Distribution-Based OTU-Calling |
| Rarefaction Curve |
| Shannon-Wiener Curve |
| Phylogenetic Tree |
| Rank Abundance Curve |
| Relative Abundance |
Heatmap |
| VENN |
| Principal Components Analysis (PCA) |
| Hierarchical Clustering |
| Community Structure |
RDA/CCA Analysis |
| Multiple Contrast |
| Network Analysis |
| Functional analysis |
BLASTX, KEGG, eggNOG, CAZy, CARD, ARDB etc. |
| PICRUSt |
| TAX4Fun |
| Custom Analysis |
Custom Analysis |
Related Sections
Services:
Solutions:
Products:
Resource:
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.





 Figure 1. High-throughput sequencing analysis process
Figure 1. High-throughput sequencing analysis process  Figure 2. PCR-DGGE analysis process
Figure 2. PCR-DGGE analysis process