What is the Gut Microbiome?
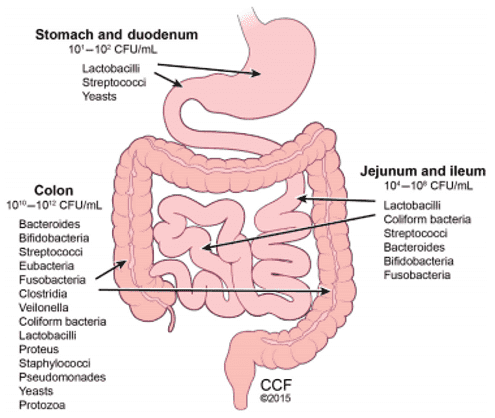
The term "gut microbiome" pertains to the composite genomics of the complex population of microorganisms located within the human gastrointestinal tract. This dynamic consortium is inclusive of diverse biological components, which encompasses bacteriophages, viruses, bacteria, protozoa, helminths (worms), and mycetes (fungi). This microbial population expands to a staggering total of almost 10^14 microorganisms, which cumulatively surpasses the total number of human body cells by an approximate ten-fold.
The cumulative weight of the gut microbiota oscillates at approximately 1.2 kilograms or possibly more, constituting a diversity range of over 1000 distinct species. Intriguingly, the quantity of genes affiliated with the human gut microbiome surges to an estimated 3.3 million, overshadowing the total count of human genes by an impressive magnitude of approximately 150 times. This staggering complexity not only underscores the biological importance of the gut microbiome but also sparks intrigue about its potential influence on human health and disease.
Gut Microbiome and Human Diseases
Within the human gut, there exist approximately 1000 to 1150 species of bacteria, which collectively amount to about a trillion, outnumbering human body cells by a factor of ten. A smaller population of fungi and viruses also co-inhabit the gut environment. The bacterial species predominantly found in the human gut are divided among two to five phyla, with proteobacteria and firmicutes asserting dominance, and actinobacteria, spirochaetes, and verrucomicrobia serving as secondary contributors. There is a profound connection between human health and gut microbiome composition, and the intricate host-microbiome interactions uphold the ecological equilibrium of the gut microbiota.
In recent years, exponential advancements in molecular biology, high-throughput sequencing technologies, and bioinformatics have set significant strides in gut microbiome research. A growing body of research corroborates that the gut microbiome is not just closely related to intestinal health, but is also associated with metabolic disorders like obesity and diabetes, neuro-psychological ailments like anxiety and depression, as well as various types of cancer. Consequently, studying the interrelation between the gut microbiome and diseases could potentially pave the way for novel therapeutic breakthroughs.
Inflammatory Bowel Disease (IBD) and the Gut Microbiome
Inflammatory bowel disease (IBD), a term referring to conditions such as Crohn's disease (CD) and ulcerative colitis (UC), involves complex interactions with the gut microbiota. The gut microbiota is generally categorized into three clusters: beneficial microbes, conditionally pathogenic microbes, and outright pathogenic microbes.
Published research has delineated that when beneficial gut microorganisms establish colonies in the mucosal layer of the intestinal epithelial cells, they generate metabolic byproducts, such as short-chain fatty acids (SCFAs). Notably, these SCFAs gain entry into the epithelial cells, act as triggers for immune inhibitory cells, and effectively inhibit the activation of inflammatory responses.
Nonetheless, under circumstances involving the proliferation of certain pathogenic microorganisms, the integrity of the mucosal barrier could be significantly breached. This compromise effectively leads to a shrinkage of the thickness of the mucosal layer and an expansion of the intercellular spaces. This augmented permeability allows pathogenic microbes and their detrimental metabolites to infiltrate the epithelial cells. The subsequent induction of inflammatory responses could progress into conditions classified under inflammatory bowel disease.
Obesity and the Gut Microbiome
In the face of an escalating global obesity epidemic, the nexus between gut microbiota and obesity has garnered heightened attention within the scientific community. Comparative research has provided evidence that both the density and diversity of gut microbiota are notably diminished in obese subjects relative to their healthy-weight counterparts. A differential distribution of microbial populations is observed in the intestines of obese individuals, designated by diminished colonies of Bifidobacterium and Firmicutes in contrast with a prevalence of Clostridium and Enterobacteriaceae. Sustained dietary patterns high in fats and sugars are seen to instigate a shift in gut microbiota composition, with conditionally pathogenic bacteria gaining dominance and beneficial, symbiotic bacteria receding. This imbalance engenders what is termed gut microbiota dysbiosis, a condition in which caloric intake is more efficiently converted into adipose tissue and stored beneath the skin, thereby accelerating obesity's onset and progression.
Gut Microbiota and Diabetes
Dysbiosis of the gut microbiome can engender various metabolic disorders and has a direct bearing on obesity and fatty liver diseases. It also constitutes a crucial risk factor for diabetes. Both type 1 and type 2 diabetes are associated with gut microbiota dysbiosis. In type 1 diabetic patients, the abundance of Bifidobacterium and Lactobacillus is negatively correlated with blood glucose levels, while that of Clostridium is positively correlated. Patients with type 2 diabetes exhibit a reduced abundance of Lactobacillus, increased abundance of Bifidobacterium, and display a positive correlation between blood glucose levels and the Proteobacteria phylum. Alterations in the normal gut microbiota directly influence the hepatic vascular barrier, altering its permeability to bacteria and their metabolic byproducts, thereby triggering an inflammatory response from the immune system. This interferes with insulin tolerance factor-induced diabetes.
Gut Microbiota and Psychiatric Disorders
Increasing preclinical and clinical evidence suggests a significant relationship between gut microbiota and the brain, emotions, and cognitive behavior. This intricate connection has been observed in psychiatric disorders such as depression, anxiety, schizophrenia, autism, Alzheimer's disease, and Parkinson's disease, where gut microbiota have been found to play a substantial role. The gut-brain axis appears to hold a crucial role in the pathological mechanisms of these diseases.
Gut Microbiome and Cancer
Disruption of the intestinal barrier, leading to alterations in the gut microbiome, has substantial implications on cancer development and progression. By modulating host metabolic pathways, immune responses, and inflammation, the altered gut microbiota can induce cancer-associated pro-inflammatory cytokines. Uncontrolled inflammation, in turn, fosters the growth of potentially harmful bacteria adapted to inflammatory environments. Certain oncogenic bacteria can transform the immune composition within the tumor microenvironment, thereby assuming a pivotal role in the onset and advancement of cancer.
Intestinal Microbiota and Disease Research Paradigms
Exploring the relationship between gut microbiota and diseases can be mainly divided into three types of research models: correlational studies, causal investigative studies, and research applying microbial interventions in diseases. These three, while independent of each other, are closely interrelated.
Disease and Microbiota Correlation Studies
① Characteristic Microbiota Studies
These studies mainly aim to objectively describe the features of the human microbiota composition, explaining the relationship between a particular disease or phenomenon and the respective commensal microbiota.
Research approach:
Relatively straightforward, this type of research typically sets up cohorts of diseased and healthy groups. By comparing large samples between the two groups, the characteristic microbial constitution of specific populations can be determined.
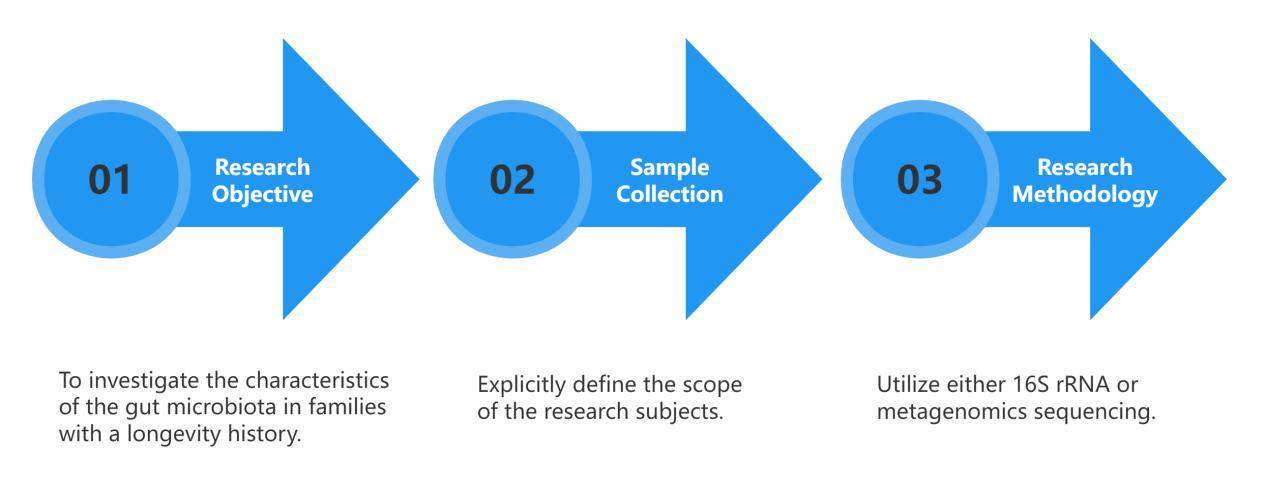
② Research on Factors Influencing Microbial Composition
Multiple concurrent factors, ranging from genetic predisposition and lifestyle choices to dietary habits, level of physical activity, and even the environmental milieu, contribute to shaping the composition of the intestinal microbiota. The aforementioned determinants play a pivotal role in maintaining the equilibrium of the gut's microbial ecosystem. In particular, research that delineates influences on the microbial makeup within neonates – elements such as delivery modalities, maternal dietary patterns during gestation, methods of infant feeding (exclusive breastfeeding, early incorporation of solid foods, formula feeding), and premature birth – bears considerable significance. Appreciating the variables that mould the neonate's gut microbiota is instrumental for subsequent health guidance and the preservation of infantile health.
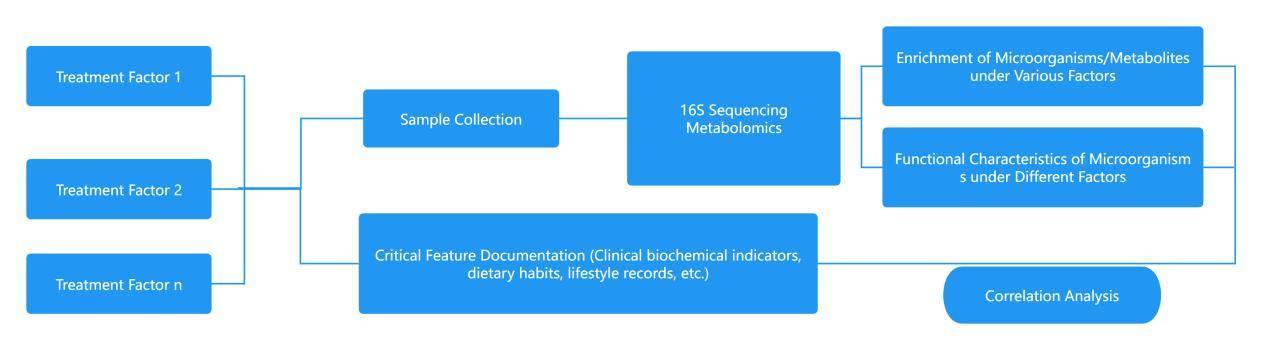
Studies on the Causal Relationships between Diseases and Microbiota
① functional verification of potential pathogenic or beneficial bacteria and research on disease mechanism:
The methodologies for functional verification of potential pathogenic or beneficial bacteria and investigation of disease mechanisms are as follows:
Identify one or more target bacteria. Carry out validation using animal models, discuss the relationship between such bacteria and the disease by analysing the clinical physicochemical indicators.
Collect and process fecal samples from animal models, sequence them, and investigate how such bacteria influence the pathogenicity or amelioration of diseases within the gut microbiota.
Synthesis of clinical indicators, physicochemical results, and microbiological results for a comprehensive analysis of the operational mechanism.
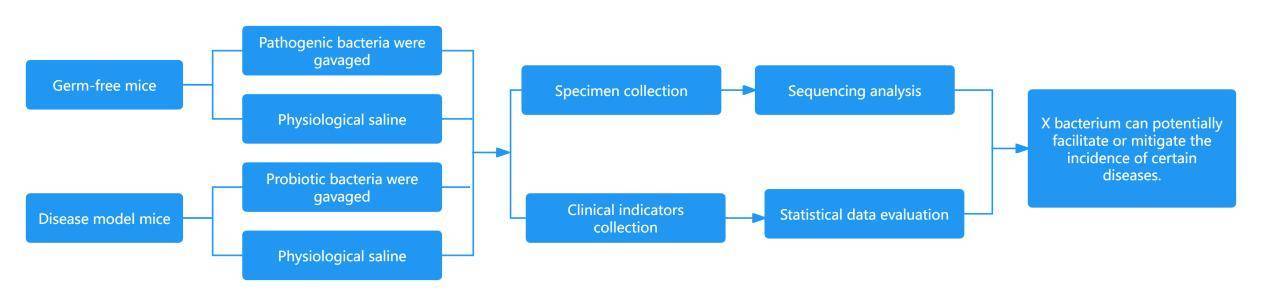
② Studies on the Relationship between Disease Progression and Microbial Communities.
Research approach:

Research on the Interventions of Microbial Communities
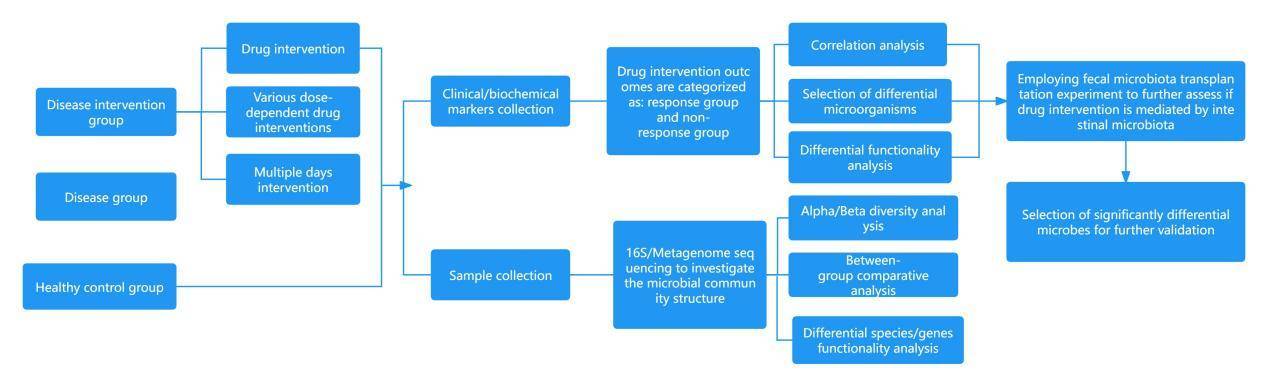
Research Outline:
Evaluate the therapeutic effects (clinical indices, biochemical indices, etc.) of various therapy methods (different drug interventions, varying dosages of the same drug, interventions over different durations, probiotics, fecal microbiota transplantation, etc.) on disease. The effectiveness of the treatment is adjudicated based on clinical indices.
Compare the differences in the compositions of the microbiota among disease group, disease intervention group, and healthy control group.
Validate whether the therapeutic method achieves its treatment effect through altering the microbial communities.
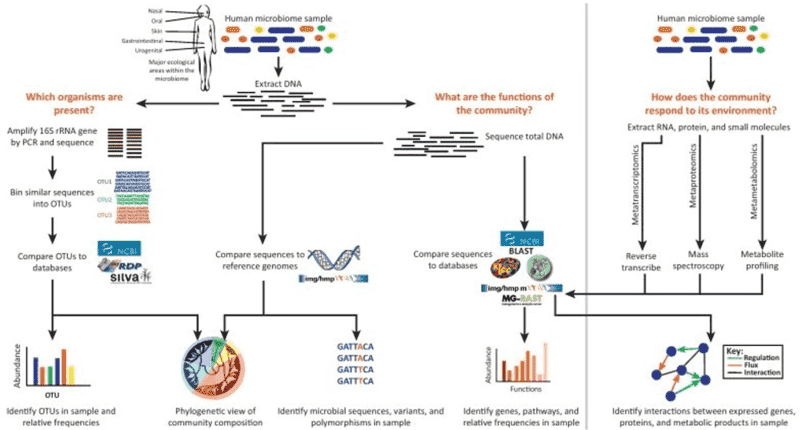
Research Methods in Gut Microbiome Studies
The primary methods currently employed for the study of the gut microbiome are 16S rRNA sequencing/genomic metagenomics and metabolomics—an approach that can be deemed as prevailing in the field. Metabolomics provides a closer reflection of phenotypes. The integration of genetic sequencing and metabolomics allows for a comprehensive elucidation of underlying mechanistic issues.
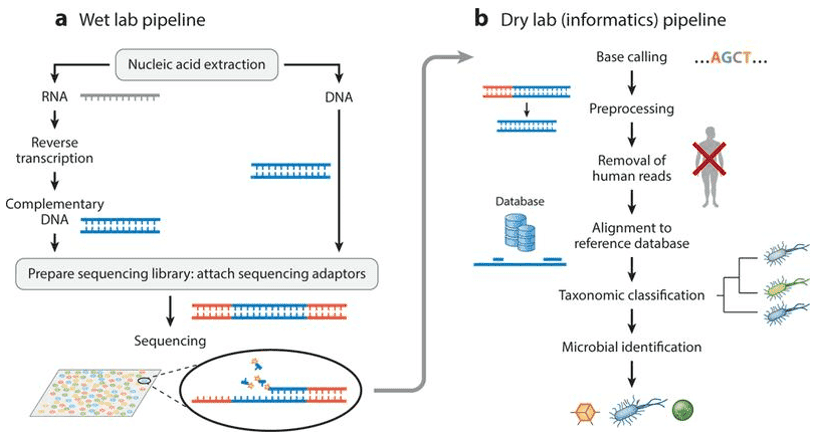
Research Methodologies in Microbiome Culturomics
Traditionally, microbiome culturomics practices involve the cultivation and isolation of microbes using various culture media, followed by identification of bacterial species through methods such as Gram staining, biochemical tests, and serological examinations. Microbial quantities are determined using methods like serial dilution and colony counting.
Rapid advancements in technology have revolutionized contemporary culturomics by stimulating the growth of hard-to-cultivate bacteria using a variety of culture conditions. In conjunction with mass spectrometry and sequencing techniques for bacterial identification, not only has the number of cultivable human bacteria increased by hundreds, but it's also been employed in the clinical separation and identification of pathogenic bacteria, leading to the discovery of new species.
High Throughput Sequencing Methods
16S rRNA gene sequencing (also known as 16S rDNA or microbiome amplicon sequencing): 16S rRNA gene sequencing involves the amplification of bacterial 16S rDNA from sample DNA via PCR. Due to sequencing read length limitations, typically only 1-2 regions (V4, V3V4, V4V5, for example) are amplified followed by high-throughput sequencing analyses. Then, based on the wealth of sequence information obtained, bacterial species are classified and identified.
16S rRNA gene sequencing offers the advantages of simple methodology and cost-effectiveness, typically being the initial choice for screening when the correlation between a disease and the gut microbiome is unclear. Consequently, this method provides fundamental data for more in-depth subsequent research.
Metagenomic Sequencing: This method eliminates the need for isolating and culturing microorganisms in the environmental samples. It essentially involves the high-throughput sequencing of the total DNA of all the microorganisms present in the environmental samples. The main applications of metagenomic sequencing include studying the genetic composition and functionality of microbial communities, decoding relative abundance and diversity of microorganisms, analyzing the relationships between microorganisms and their environment or host organisms, and exploring and studying new genes with specific capabilities.
Metagenomic Sequencing allows for simultaneous acquisition of species, gene, and functional information about gut bacteria, fungi, and viruses. It is commonly used for data mining of gut microbiome genes and functions, studying the mechanisms of diseases related to gut microbiome, or mechanisms of drug action, among other applications.
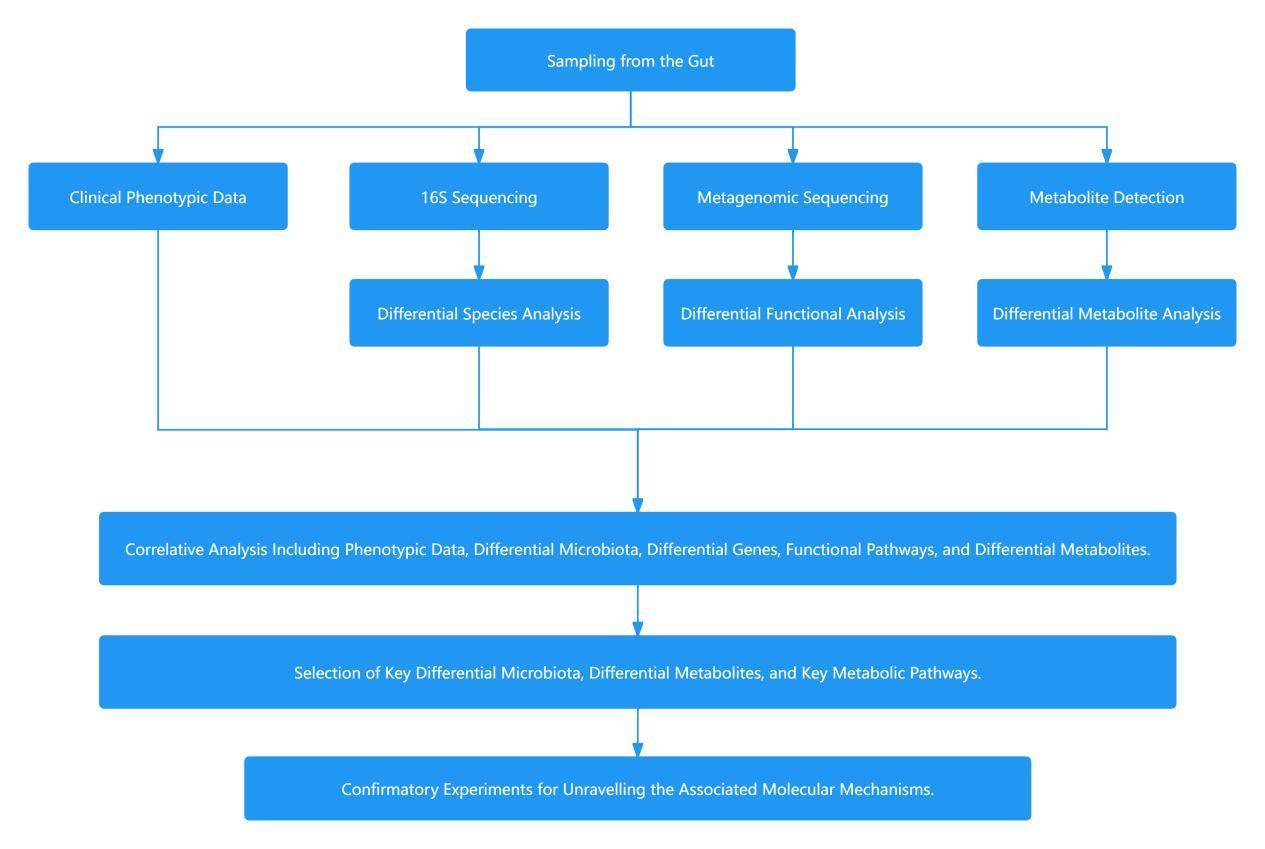
Study Approach for Gut Microbiota
References
- Dominguez-Bello M G, Godoy-Vitorino F, Knight R, et al. Role of the microbiome in human development. Gut, 2019.
- Fz A, Da B, Ji Y, et al. The gut microbiome in health, disease, and clinical applications in association with the gut bacterial microbiome assembly. The Lancet Microbe, 2022.
- Janney A, Powrie F, Mann E H. Host–microbiota maladaptation in colorectal cancer. Nature.
- Sepich-Poore G D, Zitvogel L, Straussman R, et al. The microbiome and human cancer. Science, 2021, 371(6536): eabc4552.
- Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nature Reviews Microbiology, 2020.
- Wei Miao-Yan, Shi Si, Liang Chen, et al. The microbiota and microbiome in pancreatic cancer: more influential than expected. Mol Cancer, 2019, 18: 97.
- Natalini Jake G, Singh Shivani, Segal Leopoldo N, The dynamic lung microbiome in health and disease. Nat Rev Microbiol, 2023, 21: 222-235.
- Liu Ya, Guo Yifan, Liu Zheyu, et al. Augmented temperature fluctuation aggravates muscular atrophy through the gut microbiota. Nat Commun, 2023, 14: 3494.
- Gan Lin, Feng Yanling, Du Bing, et al. Bacteriophage targeting microbiota alleviates non-alcoholic fatty liver disease induced by high alcohol-producing Klebsiella pneumoniae. Nat Commun, 2023, 14: 3215.
- Huang Yinhua, Wang Zhijie, Ye Bo, et al. Sodium butyrate ameliorates diabetic retinopathy in mice via the regulation of gut microbiota and related short-chain fatty acids. J Transl Med, 2023, 21: 451.