Scalp Microbiome
With multiple studies showing that modifications in microbial community composition are correlated with disease states as varied as irritable bowel syndrome, acne, periodontal diseases, and atopic dermatitis, it is well acknowledged that the human microbiota plays an important role in maintaining human health. A scalp situation characterized by excessive flaking of the skin, often followed by dryness and itching, is the suggested microbiome-induced cosmetic condition dandruff.
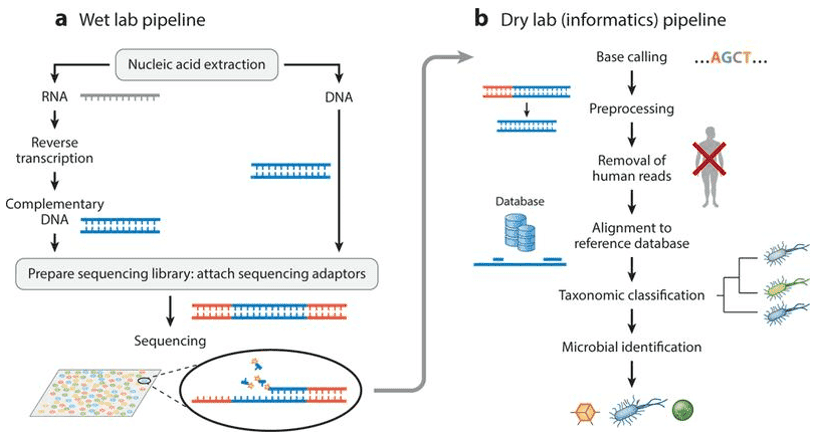
The high-performance assessment of bacterial diversity at a level not possible only a few years ago has been initiated by the latest developments in next-generation sequencing. The study outlines a meta taxonomic profiling position adopted on both healthy and dandruff scalps to evaluate the scalp microbial profile of the resident bacterial and fungal communities. The relative abundance evaluation supplied by metataxonomic analysis is complemented by qPCR of the key members of the scalp microbiome bacterial and fungal community.
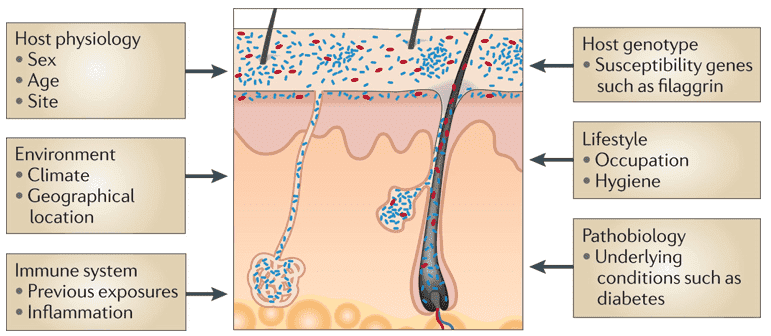
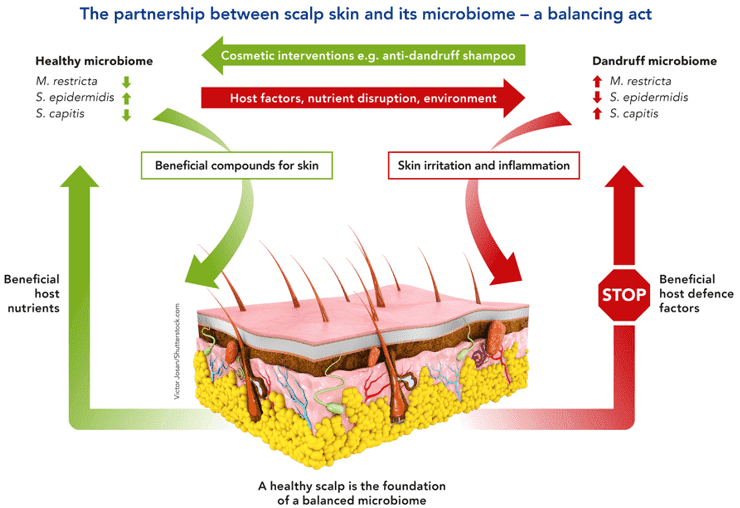 Figure 1. The partnership between scalp skin and its microbiome. (Rudden, 2020)
Figure 1. The partnership between scalp skin and its microbiome. (Rudden, 2020)
Sample Method for Analyzing Scalp Microbiome
Sample collection
Sterile phosphate-buffered saline can be employed to the sample site utilizing a sterile plastic disposable pipette. With a Teflon rod for one minute, the scalp surface is then slowly flustered. At the same sampling site, this cycle is continued and the samples are pooled. Prior to DNA extraction, specimens are put on ice.
DNA extraction
All samples are centrifugally concentrated, supernatants are discarded, and cells are resuspended in a sterile TE buffer. Cell lysis is conducted, cells suspensions are incubated with agitation preceded by bead beating. DNA is subsequently obtained from the specimens and purified.
Using general primers, bacterial and fungal-derived Polymerase Chain Reaction (PCR) amplicons are generated for each data set. A 550 bp fragment of the 16S rRNA gene for bacteria is amplified using an in-silico primer set initially analyzed using open-source software. For fungi, the fungal rRNA gene region ITS2 is also amplified.
PCR reactions are performed in triplicates for bacteria and fungi. The samples are amplified. The replicated PCR reactions are pooled, visualized by gel electrophoresis, and purified using the DNA extraction kit from each sample. Then DNA is further purified through magnetic beads. PCR amplicons have been quantified from each data set.
Statistical analyses
a. Metataxonomics
At the OTU level, sample library sizes can be first normalized. The rarefied OTU tables are further collated to the level of the genus and assessments for both are conducted. Using a Bray-Curtis dissimilarity with non-metric multidimensional scaling, sample and group beta diversity is defined. Variance analysis between scalp types and site score classes is conducted on the Bray-Curtis dissimilarity matrix using permutation analysis of variance. Calculations can be done using the Vegan, Party, and HMP R packages with q-value for false discovery control.
b. qPCR
In triplicate, the qPCR measurements are done and these are summarized using the median value. To analyze data with independent factors for each species, mixed models are utilized. As fixed effects, TWHS/site grades are included and the subject is added as a random effect.
References
- Rudden M, Herman R, Rose M, et al. The molecular basis of thioalcohol production in human body odour. Scientific reports. 2020 Jul 27;10(1).
- Grimshaw SG, Smith AM, Arnold DS, et al. The diversity and abundance of fungi and bacteria on the healthy and dandruff affected human scalp. PloS one. 2019 Dec 18;14(12).
- Saxena R, Mittal P, Clavaud C, et al. Comparison of healthy and dandruff scalp microbiome reveals the role of commensals in scalp health. Frontiers in cellular and infection microbiology. 2018 Oct 4;8.


 Figure 1. The partnership between scalp skin and its microbiome. (Rudden, 2020)
Figure 1. The partnership between scalp skin and its microbiome. (Rudden, 2020)