The Importance of Microbes in the Air
Microorganisms such as bacteria, virus, and fungi am teeming in the air. These biological particles contributes to the ecological balance and play an important role in agriculture, the biosphere, cloud formation, global climate, and atmospheric dynamics. However, they also affect the environmental equality, and even act as a medium for the spread that leads to air pollution and human diseases. Airborne microorganisms are a major cause of human respiratory diseases, such as asthma, allergies, and pathogenic infections. Both cultivation-dependent and –independent methods can be used to explore microbial diversity in the air, uncovering the potential impacts on the natural environment and human disease.
 Request a Quote
Request a Quote
What Can We Do?
- 1. Measuring the types and abundance of microorganisms in the samples;
- 2. Qualitative and quantitative analysis of certain microorganisms in the samples;
- 3. Obtaining the microorganism strain by anaerobic culture.
Note: Our service is for research use only, and not for therapeutic or diagnostic use.
Detectable Objects
The various microbiome samples from indoor or outdoor environments. We have ever analyzed various samples collected from hospitals, schools, industrial buildings, etc.
Detectable Microorganisms
Bacteria, fungi, virus.
Detection Methods
Next-generation sequencing (NGS), long-read sequencing, PCR-DGGE, Real-time qPCR, anaerobic culture.
Technical Platforms
Illumina HiSeq/MiSeq, Ion PGM, PacBio SMRT systems, Nanopore systems, PCR-DGGE, Real-time qPCR, anaerobic culture.



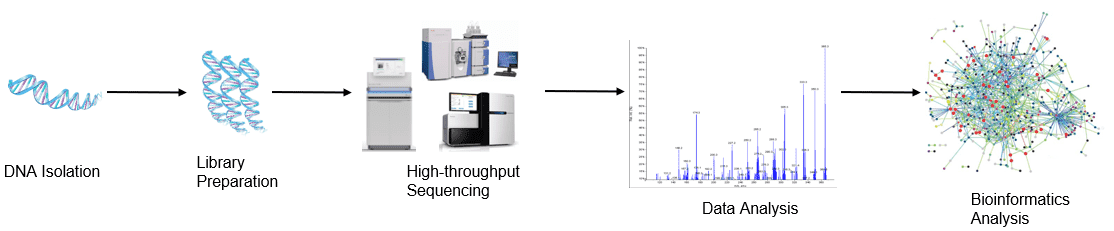 Figure 1. High-throughput sequencing analysis process
Figure 1. High-throughput sequencing analysis process Figure 2. PCR-DGGE analysis process
Figure 2. PCR-DGGE analysis process