The importance of microbial diversity analysis in extreme environments

The extreme environment such as volcanoes, glaciers, and deep seas, is an indispensable part of the natural environment. Its condition is too harsh for human, plants, and animals to exist. However, there is probably a large number of microorganisms living in such places, which are called extremophiles. These microorganisms represent the limits to adapt to the environment and are treasures of biological genetics and functional diversity. They not only play an important role in the origin and evolution of life, but also have great applications in studying specialized genotypes, physiological mechanisms and metabolites, revealing the mechanism of these microorganisms to colonize hostile ecosystems. The enzymes isolated from them are currently being exploited for their commercial interest and industrial applications. At present, extreme microbes have become a hot field of international research.
 Request a Quote
Request a Quote
Accelerate microbial research in extreme environments
With the development of next-generation sequencing and bioinformatics tools, it is possible and much easier to conduct systematic classification, gene function analysis, and pathway analysis in extreme environments. We provide a broad array of microbial genomics solutions to help researchers explore the diversity of microbial community structures in extreme environments and maybe find resources that have great potential for industrial applications.

Microbial identification and quantification
16S ribosomal RNA (16S rRNA), the most common housekeeping genetic marker in bacteria, is ideal for bacterial detection and bacterial diversity analysis. 18S rRNA and ITS are ideal for fungal detection and diversity analysis. We provide both short-read and full-length 16S/18S/ITS amplicon sequencing and qPCR services for microbial identification and quantification in extreme environments.

Microbial diversity analysis
We utilize 16S/18S/ITS amplicon sequencing and metagenomics to help you simultaneously detect dominant species, rare species, and unknown species in your sample, and obtain the microbial community composition and relative abundance. In order to improve identification accuracy and alignment rate, we also provide long-read sequencing services utilizing PacBio SMRT sequencing or Nanopore sequencing.

Bioinformatics analysis
After sequencing, we process the data through our powerful bioinformatics analysis platform. First, the raw data is filtered out or trimmed, and the qualified data is retained for alignment, assembly, taxonomic assignment, diversity analysis, phylogenetic analysis, network analysis, correlation analysis, functional prediction and so on.
The main research direction of extremophiles:
- Discovery of new microbial species.
- Discovery of new genes, enzymes, or metabolites that have application potential.
- Microbial function analysis and gene function analysis.
- The molecular mechanism of microbial adaptation.
- Microbial evolution studies.
What can we do?
- 1. Identification of extremophiles
- 2. Quantitation of extremophiles
- 3. Studying the evolution and function of extremophiles, and the relationship between microorganisms and the environment
Note: Our service is for research use only, and not for therapeutic or diagnostic use.
Detectable Objects
Bacteria, eukaryotes, archaebacteria.
Detection Methods
Next-generation sequencing, third-generation sequencing, PCR denaturing gradient gel analysis (PCR-DGGE), real-time PCR
Technical Platforms
Illumina HiSeq/MiSeq, Ion PGM, PacBio sequencing, Nanopore sequencing, PCR-DGGE, Real-time qPCR, clone library etc.
Sample Requirement

DNA ≥ 300 ng, 1.8<OD260/280<2.2, concentration ≥ 10 ng/μl, Volume ≥ 10 μl.
Ensure that the DNA is not degraded or slightly degraded. Avoid repeated freezing and thawing during sample storage and shipment.
Please use enough dry ice or ice packs during shipment.
Nucleic acid samples should be stored at -20 °C.
Workflow
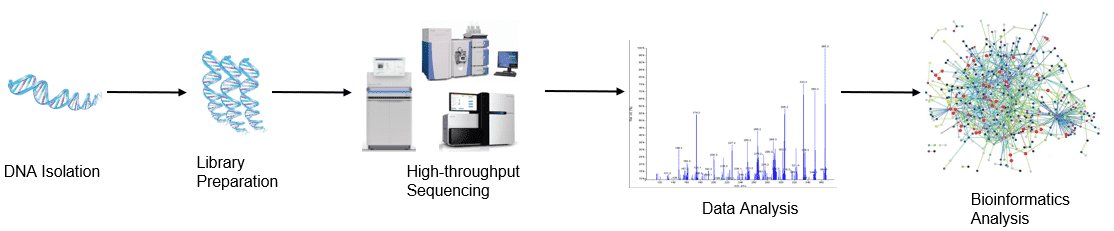 Figure 1. High-throughput sequencing analysis process
Figure 1. High-throughput sequencing analysis process
 Figure 2. PCR-DGGE analysis process
Figure 2. PCR-DGGE analysis process
Bioinformatics Analysis
| Basic Analysis |
Routine Analysis (According to Customer Requirements) |
Advanced Data Analysis |
| Sequence Filtering and Trimming |
Heatmap |
Phylogenetic Tree |
| Sequence Length Distribution |
VENN |
LDA-Effect Size (LEfSe) |
| OTU Clustering and Species Annotation |
Principal Components Analysis (PCA) |
Network Analysis |
| Diversity Index |
Microbial Community Structure Analysis |
Correlation Analysis |
| Shannon-Wiener Curve |
α Diversity Index Analysis |
|
| Rank-Abundance Curve |
Matastats Analysis |
|
| Rarefraction Curve |
Weighted Unifrac test |
|
| Multiple Contrast |
CCA/RDA Analysis |
|
| Heatmap |
|
|
| Principal Components Analysis (PCA) |
|
|







 Figure 1. High-throughput sequencing analysis process
Figure 1. High-throughput sequencing analysis process
 Figure 2. PCR-DGGE analysis process
Figure 2. PCR-DGGE analysis process