With the advancement of science and technology, human needs, especially in high-income countries, are becoming more specific and personalized. Likewise, maintaining and addressing issues in health is at the forefront of everyone's concern. Thus, with the emerging technology and diagnostic techniques available today, when taking a holistic approach to health and research and development, one area of interest that has recently become a buzzword in the scientific community is the study of the human microbiome, thereby birthing the Human Microbiome Project.
Microbial cells make up more than human cells in the body- an estimate of one hundred trillion, to be exact! – that scientists are still figuring out the exact roles of these communities of microorganisms found in the human, with very specific niches. These communities of microorganisms that have specialized in specific areas of the human body as commensals are what we call 'microbiome'. Of particular interest in the study of microbiomes is the host and microbiome interaction, as it has been observed that profiling microbiomes have been indicative of the health status of humans from which these microbiomes have been taken from, and insightful in the study of host-pathogen interaction. This is due to the fact that microbiomes are easily subjected to change in response to external influences and disease states, that even adjacent microbiomes have varying compositions.
So diverse are the microorganisms present in the small enclosed space of a mouth, that a dedicated database of 16S RNA gene sequences of oral phylotypes is curated and made available to the public- the Human Oral Microbiome Database. To date, the phyla of the oral microbiome which has generated the greatest number of genomes sequenced are the following, in order of greatest to least: Firmicutes, Proteobacteria, Actinobacteria, Bacteriodetes, and Fusobacteria. Data from genomes of these bacteria were substantially supplemented by the Human Microbiome Project, which has collated studies on microbiomes sampled from different parts of the human body. In the mouth, samples were taken from saliva, throat, buccal mucosa, tongue dorsum, hard palate, keratinized gingivae, palatine tonsils, subgingival plaque, and supragingival plaque.These samples then underwent high-throughput analysis using genetic sequences of 16S ribosomal RNA, which is used in genetic microbiome assays as these sequences are highly conserved.
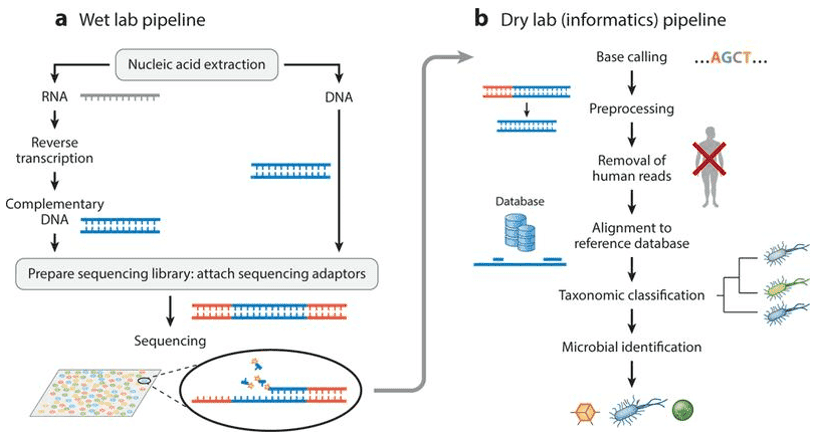
For the profiling of microbiomes, specifically oral microbiomes, researchers used 16S rRNA gene pyrosequencing and the Human Oral Microbe Identification Microarray (HOMIM), the former of which amplifies a 16S rRNA gene with the use of PCR primers to highly conserved regions, then using DNA pyrosequencing which yields a large number of shorter reads with more depth for a cheaper price. HOMIM, on the other hand, uses probes specialized to detect the usual suspect oral bacterial species, yielding a normalized data analysis. Though cheap, both methods are not without faults, which brings us to the use of shotgun sequencing in the metagenomic analysis. This method sequences genomic DNA from an environmental sample and enables the researcher to identify and characterize genes present. Further, a more advanced analysis of DNA sequences can be done, like categorizing genes identified in the community and inferring its function in the metabolic pathway. The most recent, high-throughput version of metagenomic sequencing is the use of Next Generation Sequencing, which, through bioinformatic analysis, can yield fragments of DNA to be compared to the human reference genome and used depending on the objectives of the study being conducted.
One such goal of studies being conducted on the oral microbiome is the relationship of the persisting microbial flora present in the mouth to the pathogenesis of periodontal diseases. In a study conducted by Liu and colleagues (2012) through the use of shotgun metagenomic sequencing, they found that in individuals with periodontal disease, the microbiome composition shifted from gram-positive dominated to gram-negative. In fact, there is more taxonomic diversity in those with the disease, as compared to healthy individuals where oral microbiome diversity is low, but its composition varies across humans without periodontal disease. To be fair, though, the microbiome observed in disease patients does not indicate that it the primary cause for the development of pathogenesis, or merely a by-product of the disease process. What is for certain, though, is that there is still a plethora of testable hypothesis to be studied, and discoveries to be made, more so with the advent of emerging technology in sequencing and identification and characterization of microbiome function in human health.
References
- Ahn J, Chen CY, Hayes RB. Oral microbiome and oral and gastrointestinal cancer risk. Cancer Causes Control. 2012, 23: 399-404.
- Eren AM, Borisy GG, Huse SM, et al. Oligotyping analysis of the human oral microbiome. PNAS Plus. 2014.
- Liu B, Faller LL, Klitgord N, et al. Deep Sequencing of the Oral Microbiome Reveals Signatures of Periodontal Disease. PLoS ONE. 2012, 7(6): e37919. DOI.
- Malla MA, Dubey A, Kumar A, et al. Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment. Front Immunol. 2019, 9 (2868).