For the efficient antimicrobial treatment of infected patients, the perseverance of the antimicrobial susceptibility of a clinical isolate is often essential. With developing resistance and the beginnings of multidrug-resistant microorganisms, this need is only growing. Not only therapy but also monitoring the expansion of resistant organisms or resistance genes all throughout the hospital and community needs research. To deduce therapeutic outcomes both in time and at various geographical locations, proper protocols and breakpoints have been described. The existence of a resistance gene in some cases is highly predictive of the clinical outcome of antimicrobial treatment.
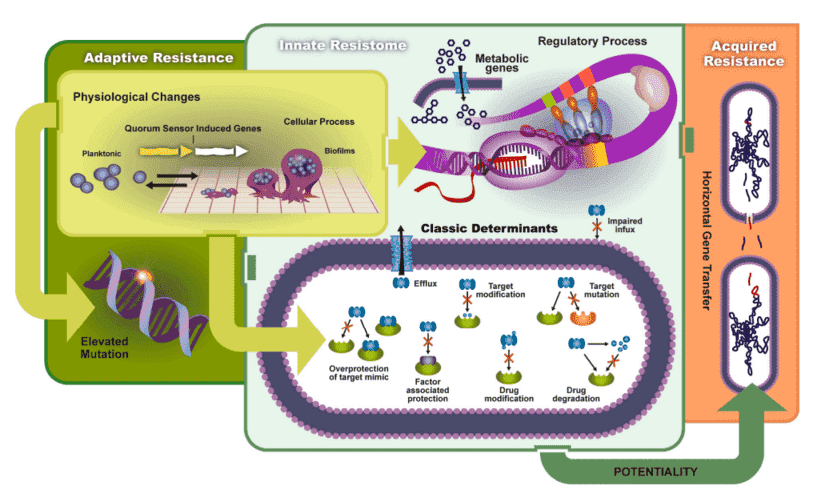
Several factors of mechanisms resulting in resistance:
1. The existence of an enzyme that makes the antimicrobial agent inactive;
2. The existence of a substitute enzyme that is hindered by the antimicrobial agent for the enzyme;
3. A mutation in the target of the antimicrobial agent decreases the antimicrobial agent's binding;
4. Posttranscriptional or posttranslational alteration of the target of the antimicrobial agent, reducing the adhesion of the antimicrobial agent;
5. Decreased absorption of the antimicrobial agent;
6. An antimicrobial agent's active efflux;
7. Excessive production of the antimicrobial agent's target.
About Polymerase Chain Reaction
Polymerase chain reaction (PCR) includes cycles of heating the mixture by a thermostable DNA polymerase for denaturing, annealing the primers, and stretching the primers. In theory, the quantity of DNA target molecules is doubled in each amplification round, but the process is rarely 100 % accurate attributed to the prevalence of inhibitors, and DNA polymerase may be constrained in subsequent amplification rounds.
Even so, recent technologies in labeling technology have widened the relevance of PCR over the last few years. The use of 5'-fluorescence-labeled oligonucleotides that were obstructed at their 3' ends was one such growth, thus stopping elongation by DNA polymerase. In addition to this unique oligonucleotide, PCR has two conventional oligonucleotides that act as primers and are chosen for the specific oligonucleotide in regions flanking it. Throughout primer extension, the special oligonucleotide hybridizes with the target and is eliminated by the 5' to 3' exonuclease activity of Taq DNA polymerase, leading to enhanced fluorescence that can be easily identified.
The creation of molecular beacons is another innovation. Molecular beacons are practically hairpin-shaped oligonucleotide probes with a fluorophore connected and a molecule that, when it is next to the fluorophore, satisfies this fluorescence. The fluorophore and quenching molecule is spatially split upon hybridization with the target and fluorescence is feasible. The usage of fluorophores with various emission spectra, in principle, allows many targets to be discriminated against. The addition of molecular beacons to PCR amplification allows for amplification monitoring in real-time. In addition, it enables a relatively simple quantitative PCR.
Detection of Antibiotic Resistance Gene
Rifampin Resistance
One of the first assays for rifampin resistance identification using PCR-SSCP has been released. In the second paper, both in a manual format with radioactively labeled amplification products and with 5' fluorescein-labeled primers for identification on an automated DNA sequencer, the assay was more intensively studied. Assessment of the research indicated that it was possible to identify all 17 of the then known rpoB gene mutations leading to resistance. Crucially valuable, the tests could be implemented to minimally grown cultures in the medium of Bactec 12B with a growth index of ≤ 100 or on sputum with a magnification of x250, with at least 10 organisms per sector. The capability of PCR-SSCP as a powerful technique for the early treatment of antimicrobial drug resistance in M. tuberculosis was clearly established. The implementation of the PCR-SSCP identification of rifampin resistance to cerebrospinal fluid specimens from patients with central nervous system tuberculosis also generated outstanding results.
Isoniazid Resistance
For the identification of katG-related isoniazid resistance, PCR-SSCP was also utilized. However, a big issue is that the mutations causing isoniazid resistance are spread over a much longer sequence, compared to rifampin resistance resulting from mutations in the rpoB gene. This requires the evaluation of complex amplification fragments, along with the fact that PCR-SSCP is most exclusionary when the amplified fragments are less than 400 bp. In relation, there is an inadequacy of all or part of the katG gene, meaning that proper measures are needed to differentiate the absence of PCR suppression from amplification products. Resistance was initially thought to be generated by complete removal of the katG gene, but with the use of PCR and PCR-SSCP, it was shown that resistance could result in both removal and mutation of the katG gene.
Multidrug Resistance
In the traditional alternative treatments for M tuberculosis, the innovation of multidrug resistance has prompted the consideration of alternative antimicrobial agents, including fluoroquinolones. However, quinolone resistance was found among clinical isolates and was easily stimulated at 2 μg/ml with ciprofloxacin (frequency, 1 in 107 to 108), although no resistance induction was noticed at higher amounts with ciprofloxacin. Therefore, in order to differentiate fluoroquinolone-susceptible from fluoroquinolone-resistant isolates, PCR-SSCP was assessed. The findings showed that discrimination was feasible between these isolates.
Isolates often display multidrug resistance, and the number of potential mutations associated implies that there can be quite a high proportion of assays required to cover them all. Several PCR tests have therefore been established that do not directly identify the presence or absence of genes and mutations that cause resistance, but either define multi-resistant strains with other properties or manage the effect of chemotherapy.
References
- Allen HK, Donato J, Wang HH, et al. Call of the wild: antibiotic resistance genes in natural environments. Nature Reviews Microbiology. 2010, 8(4).
- Rahube TO, Marti R, Scott A, et al. Persistence of antibiotic resistance and plasmid-associated genes in soil following application of sewage sludge and abundance on vegetables at harvest. Canadian Journal of Microbiology. 2016 62(7).
- Fluit AC, Visser MR, Schmitz FJ. Molecular detection of antimicrobial resistance. Clinical microbiology reviews. 2001, 14(4).
Related Sections
Services: