We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy


We are dedicated to providing outstanding customer service and being reachable at all times.
Overview of Long-read Sequencing
At a glance:
- Overview
- What Is Long-read Sequencing?
- Advantages of Long-read Sequencing
- Applications of Long-read Sequencing
- Opportunities and Challenges in Long-Read Sequencing Data Analysis
Overview
Long-read sequencing has emerged as a breakthrough method in the field of genomics, providing unrivaled insights into the complexity of genomes. While traditional short-read methods have proven limited in some cases, long-read sequencing has paved the way for a more comprehensive understanding of genetic structure and function. These technologies can generate sequences ranging from 10 kb to several megabases, ensuring more extensive, uninterrupted analysis of DNA samples.
| Pacific Biosciences (PacBio) SMRT Sequencing | Oxford Nanopore Technology (ONT) Sequencing |
| SMRTbell Template: At the heart of PacBio sequencing is the unique SMRTbell template, a topologically circular DNA molecule. The molecule is composed of a double-stranded DNA insertion fragment with a single-stranded hairpin connector at each end. Notably, the length of the DNA insert fragments can vary, even exceeding 100 KB. | Linear DNA Sequencing: Unlike PacBio's circular approach, ONT utilizes linear DNA molecules. They can range in length from a few thousand bases to several trillion bases. |
| Sequencing Mechanism: Once bound to a DNA polymerase, the SMRTbell is ready for sequencing. Here, fluorescently labeled nucleotides are doped during DNA synthesis. Each nucleotide's fluorophore emits a unique light that is captured by the camera, which then recognizes the identity of the nucleotide. With such precision, PacBio can produce reads tens of kilobases long, a significant advantage over other technologies such as Illumina. | Sequencing Mechanism: ONT sequencing utilizes motor proteins and nanopores. When the motor protein unwinds the DNA, it is driven by an electric current through the nanopore. The subsequent disruption of the current caused by the DNA translocation helps to decipher the DNA sequence. ONT has been reported to generate reads in excess of 1Mb in length, with records approaching 2.3Mb when shorter reads are combined. |
| PacBio HiFi Sequencing Technology: By targeting longer DNA (or RNA) fragments harvested directly from biological samples, it transcends the limitations faced by most short-read-length sequencing methods, which require synthetic replication of extracted DNA prior to analysis. |
The capabilities of long-read sequencing technologies such as PacBio and ONT have greatly advanced genomic research. Their ability to span larger genomic regions means that complex regions, especially repeat-dense regions, can now be sequenced more efficiently. Researchers can now study structural variation, haplotypes, and other complex genomic features in greater depth. For example, in the field of medical genomics, the comprehensive coverage of long-read sequencing is very useful. It can detect previously unrecognized genetic variants, which is critical for diagnosing rare genetic diseases. Additionally, in evolutionary biology, these technologies can help construct more accurate phylogenies by providing extended contiguous sequences.
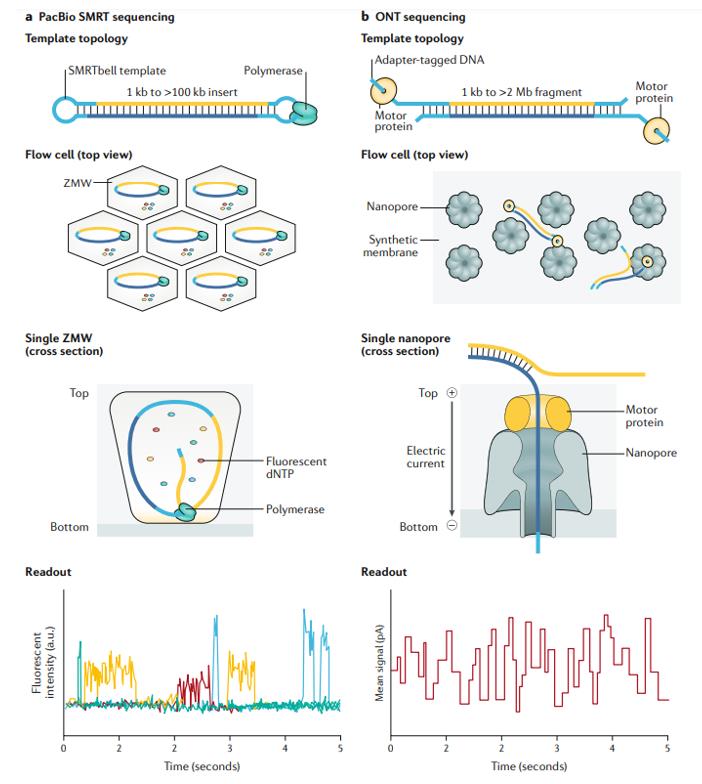 Overview of long-read sequencing technologies. (Logsdon et al., 2020)
Overview of long-read sequencing technologies. (Logsdon et al., 2020)
What Is Long-read Sequencing?
Long-read sequencing, colloquially and more technically known as third-generation sequencing, stands at the forefront of contemporary genomics, offering unparalleled depth and resolution in the deciphering of nucleotide sequences. Rather than the conventional approach of generating reads spanning a few hundred nucleotides, this avant-garde technology extends its sequencing ambit, capturing individual reads that emanate directly from single molecules, traversing expanses of thousands or, in remarkable instances, millions of nucleotides.
Contrasting its predecessor methodologies, namely the short-read sequencing which typically encompasses fragments ranging from a mere 50 to 300 base pairs, long-read sequencing exhibits its prowess by efficiently processing DNA fragments that span a substantial length, ranging from 1,000 up to an astounding 20,000 bases or even more. It's worth emphasizing that these extensive fragments are often procured in their native form - unaltered, unfragmented, and unamplified - sourced directly from the biological samples in focus. Such a methodology not only augments the accuracy and integrity of the sequencing process but also captures the nuances, intricacies, and complexities innate to the genomic structures under investigation.
Advantages of Long-read Sequencing
Comprehensive Genome Reconstruction
A key advantage of long-read sequencing is its proficiency in genome assembly. Compared to short-read methods, long-read sequencing is similar to reconstructing a detailed narrative using synthesized paragraphs rather than fragmented sentences. While short-read sequences may pose a challenge in assembling highly accurate genomes due to their limited length, long-read sequences provide sufficient contextual information to simplify the reconstruction process.
Accuracy and Context
Not all long-read methods are equal in terms of performance. Accuracy is the main factor that differentiates the various long-read techniques. While longer reads provide a richer context for genomic data, the essence of this advantage can be lost if they are not accurate. PacBio HiFi sequencing, for example, provides the length and accuracy to ensure that genomic data is not only comprehensive but also reliable.
Managing Repeated Sequences
When it comes to sequences that contain repeats or copy number variations, long-read sequencing offers a distinct advantage. Sequencing technologies such as Sanger sequencing and many next-generation sequencing methods often struggle with these repetitive sequences, resulting in inaccuracies. With long-read sequencing, these challenges are effectively addressed, making the technology invaluable for diagnosing diseases such as Huntington's disease, where duplication of specific sequences is a central concern.
Applications of Long-read Sequencing
Disease Research and Diagnosis
Long-read sequencing, as a technological advancement in genomic research, has served as a cornerstone for pivotal scientific breakthroughs, extending beyond mere nucleotide reading to a holistic understanding of genome structure and nuances. One such groundbreaking discovery in 2018, propelled by this technology, was the detailed mapping of the human Y chromosome's mitotic granules, intricate structures that underlie the chromosomal integrity. These mitotic granules are fundamental orchestrators of cell division, and deviations in their regular patterns have implications in chromosomal anomalies, notably trisomy 21, colloquially known as Down syndrome.
Furthermore, in neurodegenerative disorders, such as Huntington's disease, the pathological hallmarks often lie in the intricacies-specifically repetitive nucleotide expansions. The precise delineation of these DNA repeats, achievable through long-read sequencing, paves the way for not just a conclusive diagnosis but also a predictive understanding of the genetic trajectory of the disorder.
Pathogen Identification
In the rapidly evolving realm of infectious diseases, the prompt and unequivocal identification of pathogens becomes imperative, especially in clinical contexts where intervention time is paramount. Long-read sequencing, especially the nanopore variant, emerges as a formidable contender in pathogen detection. Its application was evinced during the Ebola epidemic, wherein its efficacy was demonstrated by the swift and unambiguous detection of viral RNA signatures in human blood samples, facilitating early interventions.
Genome Mapping
The genome, in its vast expanse, conceals layers of complexity that have often been elusive to traditional sequencing methods. However, long-read sequencing, with its capacity to decipher extended stretches of DNA with pinpoint accuracy, unravels the labyrinthine architecture of the genome. This granularity of insight equips researchers with a panoramic view of the genome, revealing the minutiae of genetic functionalities and potential mutations, thereby pushing the frontiers of genomic medicine and research.
Opportunities and Challenges in Long-Read Sequencing Data Analysis
Opportunities in Long-Read Sequencing Data Analysis
Deeper Understanding of Structural Variants (SVs)
Historically understudied due to technical limitations of short-read sequencing, structural variants are now within reach. Long-read sequencing can identify insertions, deletions, duplications, inversions, and translocations affecting ≥50 bp. This discovery has significant implications for understanding genetic diversity and human health.
Understanding Base Modifications
In addition to canonical DNA bases, there are modified bases that play key roles in organisms. The ability of the long-read platform to detect these modifications, such as 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC), at the particle size level is groundbreaking. This not only adds another layer to our understanding of gene regulation but also has potential for disease diagnosis and treatment.
Analysing Long-read Transcriptomics
Long-read sequencing allows the sequencing of full-length transcripts, providing a comprehensive view of variable splicing events, isoform diversity, and post-transcriptional modifications. These insights are critical to deciphering the complex language of gene expression regulation in health and disease.
Integration with Other TechnologiesOne of the advantages of long-read sequencing is its compatibility with other genomic tools. Combining long-read data with technologies such as short-read sequencing or Hi-C and optical mapping can produce more accurate and detailed genome assemblies. This integrated approach is shaping the future of genomics.
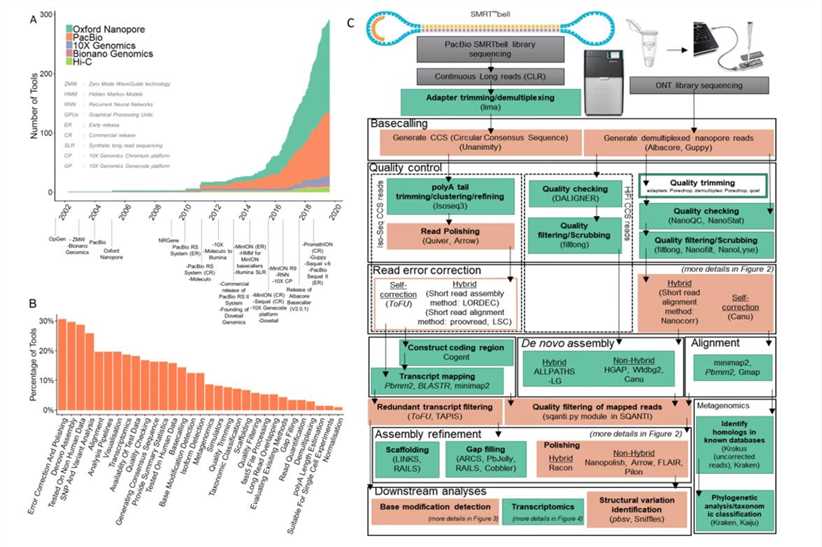 Overview of long-read analysis tools and pipelines. (Logsdon et al., 2020)
Overview of long-read analysis tools and pipelines. (Logsdon et al., 2020)
Challenges in Long-Read Sequencing Data Analysis
High Error Rates
One of the main challenges of long-read sequencing technologies (especially nanopore sequencing) is the relatively high error rate compared to short-read technologies. This requires advanced computational strategies for error correction and accurate variant identification.
Data Volume and Complexity
Long-read sequencing generates a large amount of data, posing storage, management, and processing challenges. In addition, the complexity of the data, especially when capturing rare events or processing heterogeneous samples, requires sophisticated analytical tools.
Coverage Bias
Long-read sequencing sometimes exhibits bias, especially for certain genomic regions. For example, in ONT's direct RNA sequencing protocols, coverage bias at the 3' end of transcripts may affect accurate subtype characterization.
Tool Limitations and Integration
Despite the proliferation of tools in the field tailored for the analysis of long-read data, there is still a need for better integration, standardization, and benchmarking. As technology evolves, so must the tools to ensure they remain current and able to address the latest challenges.
References
- Logsdon, Glennis A., Mitchell R. Vollger, and Evan E. Eichler. "Long-read human genome sequencing and its applications." Nature Reviews Genetics. 21.10 (2020): 597-614.
- Amarasinghe, Shanika L., et al. "Opportunities and challenges in long-read sequencing data analysis." Genome biology. 21.1 (2020): 1-16.
Related Services
PacBio SMRT Sequencing Technology
Oxford Nanopore Sequencing Technology
Transcriptomics with Long-Read Sequencing
Epigenetics and Methylation Analysis Using Long-Read Sequencing
Human Genomics with Long-Read Sequencing
Whole-Genome Resequencing with Long-Read Sequencing
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment