

We are dedicated to providing outstanding customer service and being reachable at all times.
Whole-Genome Resequencing with Long-Read Sequencing

Whole-genome resequencing focuses on the analysis of single nucleotide variants (SNV) and small fragment insertion-deletion mutations (InDel), the detection of variants in some repetitive sequence regions, and the detection of structural variants (SV) in large segments. Improvements in the accuracy and contiguity of genome assembly will inevitably enhance our understanding of more complex forms of genetic variation, which in turn will improve our understanding of mutation and evolutionary processes.
Long-read sequencing technology can span repeated elements or repetitive regions, greatly facilitating de novo assembly and SV calling. CD Genomics is a leading global life sciences company. We specialize in innovative applications of long-read sequencing technologies, genomics analysis methods, and internet concepts in the pharmaceutical and biotechnology industries, science, and agricultural research. We are now providing whole-genome resequencing services to evaluate the full DNA content of an organism, including human, plant, animal, and microbe.
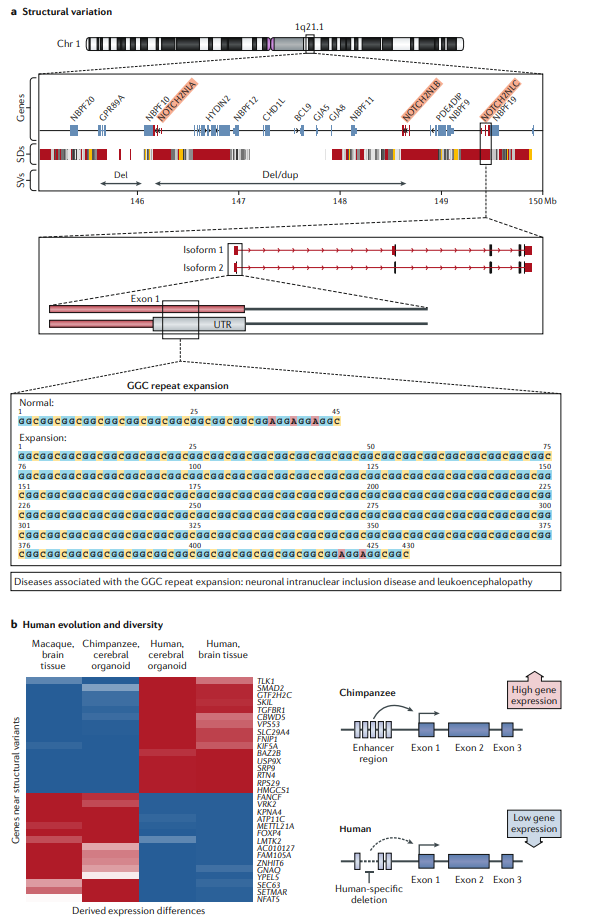 Fig1. Long-read data provide insights into the biological relevance of structural variation and human evolution and diversity. (Logsdon, G. A., et al., 2020)
Fig1. Long-read data provide insights into the biological relevance of structural variation and human evolution and diversity. (Logsdon, G. A., et al., 2020)
Advantages of Genome Sequencing with Long-Read Sequencing
- The use of long-reads (>10 kb on average) from a single DNA molecule.
- Real-time sequencing and analysis with high precision and recall.
- No PCR amplification is required, simplifying operational processes and avoiding any PCR-related bias.
- Allow direct sequencing of repeat expansions and regions with extreme GC-content.
Service Offering at CD Genomics
- Variant Calling
Variant calling refers to the sequencing and differential analysis of the genome of an individual or a species population by high-throughput sequencing technology. A large amount of genetic variation information can be obtained, such as single nucleotide polymorphism (SNP), insertion-deletion (InDel), structural variation (SV), copy number variation (CNV), and so forth. This basic information can be further used to develop molecular markers, establish a genetic polymorphism database, and provide data basis for the subsequent revelation of evolutionary relationships and mining of functional genes. - Genome-Wide Association Study (GWAS)
Genome-wide association study (GWAS) is a method to analyze the association between molecular marker data and phenotypic traits using a high density of genome-wide genetic markers for the population under study. In the past two decades, GWAS has become an effective tool for studying the genetic structure of complex traits in humans, animals, and plants. - Population Evolution
Population evolution research refers to the study of species evolution at the molecular level by obtaining information on the variation of each subgroup in a natural population of a specific species through different sequencing methods, involving biological issues such as population genetic diversity, genetic structure, gene flow, species formation mechanisms, and population dynamic evolution. - Human Whole-Genome Resequencing
We are now providing PacBio SMRT sequencing for human whole-genome sequencing. Our service enables comprehensively exploring the SNP/ InDel/ CNV/ SV variants in the genome and provides important information for screening disease causative and susceptibility genes and studying pathogenesis and genetic mechanism. - Animal/Plant Whole-Genome Resequencing
Animal and plant whole-genome resequencing (WGRS) refers to sequencing the entire genome of a plant or animal and aligning it to a known reference genome. We have two long-read sequencing platforms, namely PacBio SMRT sequencing and ONT nanopore sequencing, which provide comprehensive access to coding and non-coding regions of chromosomal, mitochondrial DNA, and chloroplast (plant) DNA. Applications we serve include, but are not limited to, studies on the origin and evolution of species, genome-wide association studies (GWAS), and agricultural breeding programs. - Microbial Whole-Genome Resequencing
Microbial whole-genome resequencing is the sequencing of the whole genome of bacteria, fungi, or other microbes, and comparing it to multiple known reference genomes. We are dedicated to providing rapid and reliable information on microbial genome sequences, detecting key and low-frequency mutations, as well as other genetic variations among microbial strains.
Thank you for your interest in our services. In order to get more details about our services, please contact us. We've got everything covered for your needs.
References
- Logsdon, G. A.,et al. (2020). "Long-read human genome sequencing and its applications." Nature Reviews Genetics, 21(10), 597-614.
- Amarasinghe, S. L., et al. (2020). "Opportunities and challenges in long-read sequencing data analysis." Genome biology, 21(1), 1-16.
- Tedersoo, L., et al. (2021). "Perspectives and benefits of high-throughput long-read sequencing in microbial ecology." Applied and Environmental Microbiology, 87(17), e00626-21.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment