CD Genomics provides fungal whole genome sequencing with the PacBio Sequel system to offer more insights into genetic structure and functions. We are best in the knowledge, practice, and experience.
The Introduction of Fungal Whole Genome de novo Sequencing
Fungi play vital roles in the biosphere and are closely related to human life with important medical and economic value. Fungi are involved in a wide range of activities—some fungi are decomposers, parasites or pathogens of other organisms, and others are beneficial partners in symbiosis with animals, plants or algae. Some fungi affect human health in various ways. Fungi are also the major sources of antibiotics, such as penicillin from the fungus Penicillium.
Obtaining fungal whole genome sequence is the basics of fungi research, and to better understand fungal biodiversity, growth, nutrition, physiology, genetics, metabolism, and ecology. Thus, hundreds of fungal genomes have been sequenced and are publically available today with a significant increase, although these initiatives have typically yielded considerably fragmented genome assemblies, they often lack large contiguous genomic regions. Many important genomic features are contained in intergenic DNA that is often missing in current genome assemblies.
To solve this problem, CD Genomics introduces the PacBio platform. This machine uses single molecule real-time (SMRT) detection technology that achieves real-time sequencing of individual polymerase molecules. SMRT detection is based on the properties of zero-mode waveguides (ZMWs), consisting of DNA polymerases bound to nanophotonic confinement structures. This technology does not require amplification of the genomic DNA, which addresses one of the major problems of second generation sequencing technologies; thus leading to the least degree of bias and longer read lengths (average >15,000 bp, some reads >100,000 bp). With the long reads of SMRT Sequencing on the PacBio System, we can generate complete de novo assemblies for fungal genomes achieve or approximate 0 gaps or N-based errors, contig N50>1 Mb, 99.999% accuracy. Additionally, we provide microbial whole genome sequencing by using next-generation sequencing.
What are the Advantages of Fungal Whole Genome de novo Sequencing
- Comprehensive revelation of genome architecture.
- Accurate determination of gene number and positioning.
- Acceleration of biological research progress.
- Abundant project experience.
- Assurance of high-quality data.
- Short turnaround time with competitive pricing advantage.
- Specialized team for biological information analysis.
What are the Application of Fungal Whole Genome de novo Sequencing
- Fungal Research: The power of fungal genome sequencing fortifies fungal research by proposing predictions of key genes and proteins, thereby enabling an understanding of their inherent functions and possible mechanisms.
- Plant Pathogenicity: Delving into the implications of fungal genome sequencing identifies specific genes of pathogenic fungi that interact with agricultural crops. This technique illuminates the evolutionary relationships with proximate species, creating opportunities for comparative genomic studies.
- Edible and Medicinal Fungi: Analysis of annotated fungal genome sequencing reveals intricate metabolic pathways and uncovers metabolic products that are beneficial to humans. Additionally, it lays vital groundwork for investigating fungal phylogenetic relationships through comparative genomics. The realm of biological control of fungal pathogens and studies into industrial yeast is further benefitted by such research.
- Ecosystem Interaction Analysis: Fungal genome sequencing can contribute greatly to discovering genes that interact with varied biotic entities, such as plants and animals. This research enhances our comprehension of the role and impact of fungi within ecosystems, facilitating exploration of the delicate networks of species interaction and their stability within such ecosystems.
- Antifungal Research: By drawing comparisons across fungal genome sequences, researchers can unveil genes and pathways associated with antifungal resistance. This provides a theoretical bedrock for the innovation of novel antifungal agents, an essential endeavour to battle fungal infections and the challenges posed by antifungal drug resistance.
- Environmental Pollution Monitoring: Fungi exhibit a high level of sensitivity towards environmental shifts, with their population composition and metabolic activity significantly influenced by environmental determinants. By harnessing the potential of genome sequencing technology, the distribution, diversity, and metabolic potentials of fungi persisting under different environmental scenarios can be efficiently monitored. This holds paramount value for pollution tracking and ecological restoration initiatives.
- Biotechnological Applications: The outcomes derived from fungal genome sequencing lend themselves to the development of biological controls against pathogenic fungi, the enhancement of industrial yeast, and a plethora of other biotechnical applications. An in-depth comprehension of the fungal genome can augment the production and quality of bio-products, thereby fostering progress in the biotechnological realm.
Fungal Whole Genome de novo Sequencing Workflow
Our highly experienced expert team executes quality management by following every procedure to ensure comprehensive and accurate results. The general workflow for fungi whole genome sequencing is outlined below.

Service Specification
Sample Requirements
|
|
Sequencing
|
|
Bioinformatics Analysis
|
Analysis Pipeline
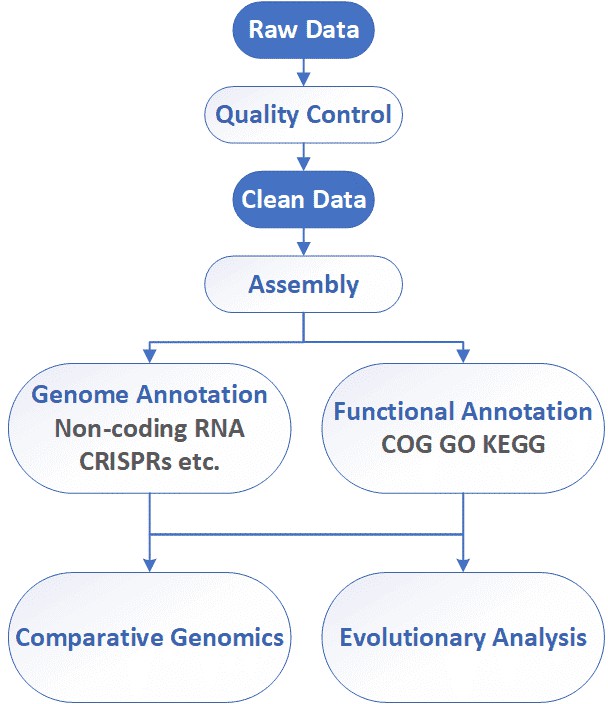
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in Fungal Whole Genome de novo Sequencing for your writing (customization)
PacBio, the third-generation sequencers, is highly robust and cost-effective and should be the platform of choice in sequencing fungi genomes, particularly for those that are well-known to be difficult-to-sequence. CD Genomics has extensive experience to offer the fungi whole genome sequencing service. Please contact us for more information and a detailed quote.
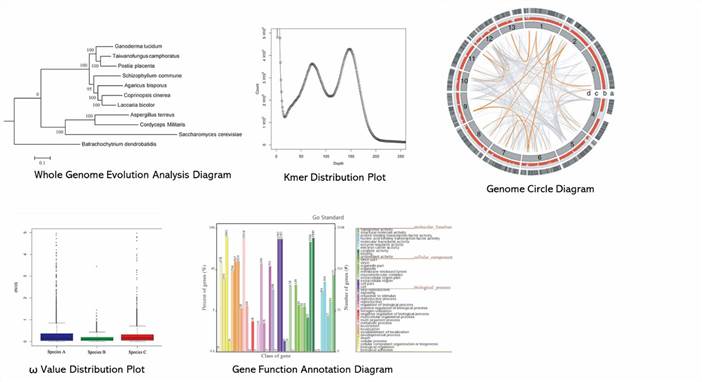
1. What are the challenges in fungal sample preparation?
Some samples are prone to degrade, so we suggest a quick library construction and sequencing after DNA extraction. Some fungal strains are difficult to culture, so library construction with a small quantity of DNA or whole-genome amplification is recommended. For samples that are difficult to isolate DNA, you can take methods from literature.
2. Why does the survey of fungal genome is required?
Compared to bacteria, fungal genomes, plasmids, and sample preparation are more complicated. Additionally, only a few fungal genomes have been published. Through genome survey, we can obtain information of genome size, GC content, repetitive sequences, and plasmids, which can pave the way to the substantial sequencing and genome assembly.
Genome Sequence, Assembly and Characterization of Two Metschnikowia fructicola Strains Used as Biocontrol Agents of Postharvest Diseases
Journal: Frontiers in Microbiology
Impact factor: 6.064
Published: 03 April 2018
Background
The yeast Metschnikowia fructicola was reported as an efficient biological control agent of postharvest diseases of fruits and vegetables, and it is the bases of the commercial formulated product "Shemer." Researchers assembled the whole genome sequence of two strains of M. fructicola.
Methods
- Metschnikowia fructicola, Strain 277
- Metschnikowia fructicola strain AP47
- DNA extraction
- Library preparation
- PacBio Sequencing
- Illumina MiSeq technology
- Gene Prediction and Functional Annotation
- Gene Expression Analysis
- Phylogenetic Tree
- Analysis of the Polymorphisms-Related Genes
Results
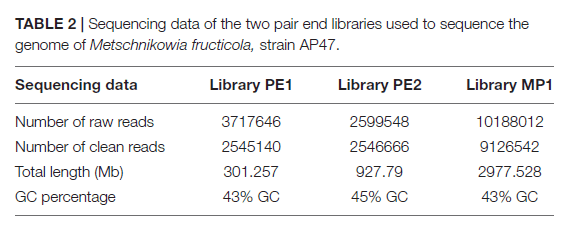
Strain 277of M. fructicola was sequenced on PacBio RS II Sequencer (P6-C4 Kit, 20 Kb Library, 24 SMRT cells, target coverage of 20X), yielded a high-quality draft genome consisting of 93 contigs with an N50 of 957,836 bp. The estimated genome size is approximately 26 Mb. Total of 8,629 genes were predicted with MAKER, and 6,262 were successfully annotated with Blast2GO and InterProScan.
M. fructicola AP47 was sequenced used by Illumina MiSeq technology, library insert size 330 bp, paired-end 300 bp. The genome size (_26 Mb) of both M. fructicola strains, as well as the rate of mutation, may suggest that M. fructicola could undergo genomic changes in order to adapt to plant surfaces, tolerate various environmental stresses and survive under restricted nutritional resources.
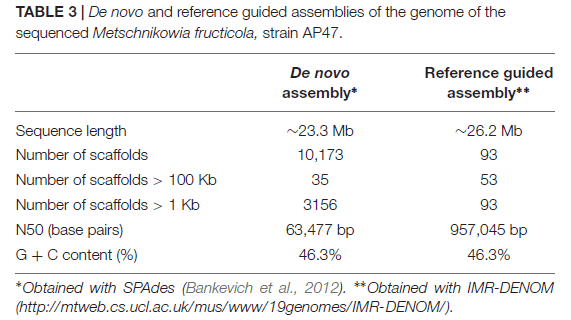
Conclusion
The study involved sequencing and comparing the genomes of two strains of M. fructicola (277 and AP47), revealing a notably high mutation rate. Further sequencing of additional strains is necessary to determine if this high mutation rate is intrinsic to M. fructicola or specific to certain geographical regions and fruit hosts. Both strains exhibited a genome size of approximately 26 Mb, indicating potential genomic adaptability to plant surfaces, environmental stresses, and limited nutritional resources. The presence of numerous secondary metabolite clusters, YAP, and CAZymes-related genes suggests M. fructicola's adaptation to the plant environment. Particularly notable was the identification of 1,145 putative CAZymes in the genome, which could serve as targets for studying enzymes for controlling fungal diseases in vivo and assessing their potential as treatments for fruits and plants.
Reference:
- Edoardo P.; et al. Genome Sequence, Assembly and Characterization of Two Metschnikowia fructicola Strains Used as Biocontrol Agents of Postharvest Diseases. Frontiers in Microbiology. 2017, 8:1-15.
Here are some publications that have been successfully published using our services or other related services:
Bacterial communities of Cassiopea in the Florida Keys share major bacterial taxa with coral microbiomes
Journal: bioRxiv
Year: 2024
Production of a Bacteriocin Like Protein PEG 446 from Clostridium tyrobutyricum NRRL B-67062
Journal: Probiotics and Antimicrobial Proteins
Year: 2024
Untangling the Role of Pathobionts from Bacteroides Species in Inflammatory Bowel Diseases
Journal: bioRxiv
Year: 2023
A chromosome-level genome resource for studying virulence mechanisms and evolution of the coffee rust pathogen Hemileia vastatrix
Journal: bioRxiv
Year: 2022
Streptomyces buecherae sp. nov., an actinomycete isolated from multiple bat species
Journal: Antonie Van Leeuwenhoek
Year: 2020
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
