Cell-Free DNA (cfdna): Definition, Differentiation, and Significance
- Cell-free DNA (cfDNA), a fragmented DNA present in the human bloodstream in a cell-free state, predominantly arises from processes of cellular necrosis and apoptosis. In particular cases, such as cancer patients, pregnant individuals, and those who have undergone organ transplantation, a distinct subset of cfDNA derived from "allogeneic" cells can be harnessed as a biomarker for genetic analysis.
- The main roles of cfDNA include: in prenatal testing, it allows for the screening of genetic diseases by detecting fetal DNA in the blood of pregnant women; in oncology, it enables the discovery of tumor-related mutations and genetic alterations by analyzing cfDNA derived from tumors, facilitating the study of tumor DNA in a non-invasive manner; in organ transplantation, it is used to monitor transplant rejection and organ health; and in cardiovascular diseases, such as myocardial infarction, the levels of cfDNA can serve as an indicator of myocardial damage.
- The significance of cfDNA applications lies in its potential as a non-invasive biomarker, making liquid biopsy techniques possible for early disease diagnosis, prognosis assessment, and therapeutic monitoring. Particularly in cancer treatment, changes in cfDNA levels are closely related to tumor burden, allowing for the monitoring of treatment response and the development of resistance. Moreover, the analysis of cfDNA can reveal tumor heterogeneity and identify multiple concurrent resistance changes in different tumor metastases, which is crucial for guiding personalized treatment and subsequent therapeutic choices.
- Technological advancements in detecting and analyzing cfDNA are swiftly progressing, with promising applications in the fields of oncology, prenatal care, and transplant medicine. As these technologies continue to evolve, cfDNA testing is anticipated to increase the precision of disease detection, refine treatment selection, and deliver more tailored healthcare solutions to patients.
What Is Cell-Free DNA (cfDNA)?
- cfDNA refers to nucleic acid fragments in the bloodstream, originating from both healthy and disease-affected cells. It is released into the circulatory system through various biological processes, including necrosis (abnormal cell death), apoptosis (programmed cell death), and direct secretion.
- cfDNA is typically found in fragments ranging from 120–220 base pairs, with a predominant fragment size of 170 base pairs, which corresponds to the length of DNA that can wrap around a nucleosome plus an additional stretch of DNA linking two nucleosome cores. The half-life of cfDNA in blood ranges from 15 minutes to 2.5 hours.
Where Does cfDNA Come From?
- cfDNA originates from various biological processes and sources within the human body. Under normal physiological conditions, cfDNA primarily comes from the degradation of genomic DNA of senescent and apoptotic cells。 When the body is affected by diseases such as cancer, trauma, organ transplant rejection, organ failure, and severe infections, abnormal necrotic cells can release a large amount of DNA into the bloodstream. Additionally, cfDNA can also be derived from the fetus-maternal unit during pregnancy, with the fetus's DNA finding its way into the maternal circulation。 Furthermore, in the context of organ transplantation, cfDNA can be released into the blood due to cell death associated with immune rejection. The release of cfDNA into the bloodstream can also occur through active secretion by cells, although the exact mechanisms are not fully understood.
- In summary, cfDNA is a complex biomarker that can be traced back to multiple sources, including cellular apoptosis, necrosis, and active secretion, making it a valuable tool in liquid biopsy and non-invasive diagnostics.
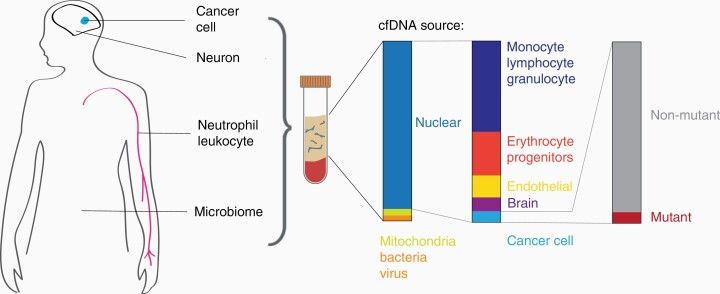 Sources of cfDNA in blood samples. (Florent Mouliere al.,2022)
Sources of cfDNA in blood samples. (Florent Mouliere al.,2022)
What Does cfDNA Test For?
cfDNA testing is a non-invasive diagnostic technique that targets and analyzes circulating DNA fragments in a patient's bodily fluids, most commonly in the blood. This testing serves several medical applications, including:
Prenatal Screening and Diagnosis
cfDNA tests, such as non-invasive prenatal testing (NIPT), analyze fetal DNA derived from maternal blood to screen for chromosomal abnormalities like Down syndrome (trisomy 21), Edwards syndrome (trisomy 18), and Patau syndrome (trisomy 13). These tests can be performed as early as the 10th week of pregnancy and provide a high rate of accuracy with minimal risk to the mother and fetus.
Oncology
In cancer management, cfDNA testing is used for early diagnosis, therapeutic evaluation, disease progression monitoring, and disease prognosis determination. Circulating tumor DNA (ctDNA), a subset of cfDNA derived from tumor cells, is particularly valuable for monitoring cancer relapse or metastasis and guiding treatment decisions.
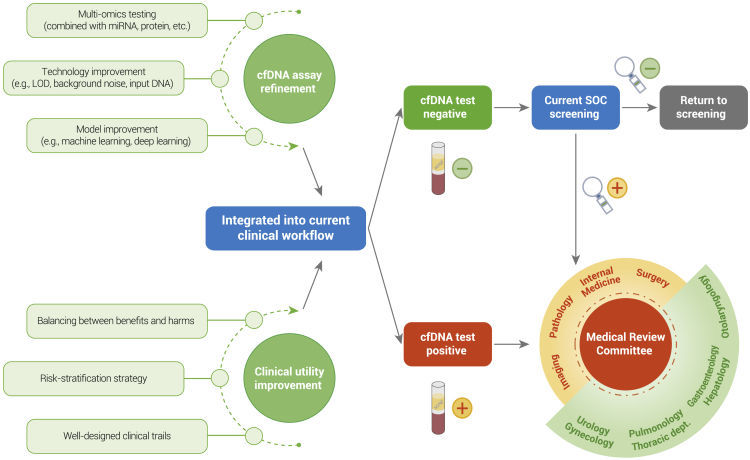 Future direction for early detection of cancers based on cfDNA tests. (Qiang Gao al.,2022)
Future direction for early detection of cancers based on cfDNA tests. (Qiang Gao al.,2022)
Organ Transplantation
Donor-derived cfDNA (dd-cfDNA) can be detected in the recipient's blood to assess allograft injury or rejection, allowing for early intervention and preventing severe outcomes.
Infectious Diseases
cfDNA can also be derived from microorganisms causing infections in humans, such as viruses, aiding in the diagnosis of infectious diseases.
Cardiovascular Disease
cfDNA has been implicated in the diagnosis of acute coronary syndrome and in predicting the severity of coronary artery lesions, with mitochondrial cfDNA (cf-mtDNA) levels elevated in cardiovascular disease patients.
In summary, cfDNA testing is a versatile tool in medicine, offering non-invasive insights into genetic conditions, cancer management, transplant outcomes, infectious diseases, and cardiovascular health.
cfDNA Methylation Analysis enables the detection and profiling of DNA methylation patterns in cell-free DNA, and CD Genomics offers the following services:
How Does cfDNA Differ From Other Types of DNA?
cfDNA differs significantly from other types of DNA in terms of origin, molecular characteristics, biological significance, and clinical applications, making it a unique biomarker that can be used for the diagnosis and monitoring of a variety of diseases.
Origin and Biological Processes
cfDNA primarily originates from DNA fragments released by cells during apoptosis and necrosis. It is DNA that is free-floating in the bloodstream, originating from both normal and diseased cells, such as cancer cells. In contrast, ctDNA is a subset of cfDNA, specifically referring to DNA that originates from tumor cells.
Molecular Size and Structure
cfDNA typically consists of shorter double-stranded DNA fragments, approximately 167 base pairs (bp) in length, and exhibits periodic fluctuations of 10.5 bp, corresponding to the pitch of nucleosome core DNA. ctDNA, on the other hand, may contain tumor-specific mutations and variations, which can be used to monitor genomic alterations and heterogeneity of tumors.
Half-Life and Clearance Rate
The half-life of cfDNA molecules in circulation is 1 hour or less, and they are rapidly cleared. This rapid clearance rate allows cfDNA to serve as a real-time biomarker for monitoring changes in disease status.
Scope of Application
The application range of cfDNA is broader than that of ctDNA. It can be used not only for cancer diagnosis and monitoring but also for prenatal screening, organ transplant monitoring, cardiovascular disease assessment, and autoimmune disease evaluation. ctDNA is mainly applied in the field of oncology for early cancer diagnosis, treatment response evaluation, and disease monitoring.
Abundance and Quality
In healthy individuals, the concentration of cfDNA is usually low, but in cancer patients, the concentration of cfDNA may increase due to the apoptosis and necrosis of tumor cells, with a portion being ctDNA released by tumor cells.
Forms of Existence
cfDNA in the bloodstream can exist in the form of free DNA fragments, DNA bound to exosomes, and DNA-macromolecular complexes. These different forms reflect the various biological processes and functions of cfDNA within the body.
What Is the Function of cfDNA?
The functions of cfDNA are diverse, and it plays a significant role in various fields such as prenatal diagnosis, organ transplant monitoring, assessment of inflammation and injury, oncology, and immune responses. With the advancement of technology, the scope and depth of cfDNA applications continue to expand.
Non-invasive prenatal diagnosis
cfDNA can be used to detect free-floating fetal DNA in the blood of pregnant women for prenatal screening of chromosomal abnormalities, such as Down syndrome (trisomy 21) and Edwards syndrome (trisomy 18).
Tumor Monitoring and Management
The application of cfDNA in oncology includes early diagnosis, determination of treatment plans, observation of therapeutic effects, analysis of metastasis risk, and monitoring of tumor recurrence. It allows for the analysis of DNA from tumors through routine blood draws, providing a minimally invasive method for molecular profiling and the ability to continuously monitor changes in tumor DNA without the need for invasive tumor biopsies.
Gene Expression Inference
The fragment characteristics of cfDNA can be used to infer gene expression, which is of great significance for understanding how genes are expressed in different tissues and how they respond to treatment.
Damage and Inflammatory Response
The concentration of cfDNA is related to the level of injury from trauma and burns. High concentrations of cfDNA are also used for predicting mortality in the ICU, as well as for predicting sepsis and septic shock, sterile inflammation, myocardial infarction, and the condition of stroke patients without radiological findings.
cfDNA Sequencing Methods
cfDNA sequencing methods play a crucial role in medical research and clinical applications, particularly in the monitoring and management of cancer. Here is a description of several professional cfDNA sequencing methods.
Target Enrichment Sequencing
This method combines hybridization capture technology to detect tumor fractions below 0.1% with high sensitivity and specificity, providing a powerful tool for disease diagnosis and longitudinal monitoring.
Targeted Error Correction Sequencing (TEC-Seq) uses molecular barcodes to distinguish true mutations from false-positive variants. Before any amplification, each DNA fragment is labeled with a unique "exogenous" DNA barcode, and paired-end sequencing read start and end genomic mapping positions are used as "endogenous" barcodes to differentiate individual molecules. This method can track approximately 30,000 sequencing reads per fragment.
Single Primer Extension (SPE)
SPE is an amplicon-based method that uses unique molecular indices to improve the accuracy of variant detection. This method uses only one gene-specific primer (GSP) to amplify each genomic region, which is less dependent on DNA fragment size compared to PCR using two primers, providing uniform coverage. Theoretically, the sensitivity threshold of this technology is 0.5-1%, with a sensitivity of over 90% and very few false positives.
Whole Genome Sequencing (WGS)
WGS is more suitable for detecting ctDNA, capable of identifying structural and non-coding variations, such as whole-genome copy number alterations, methylation profiles, and fragmentation patterns.
Plasma-Seq is a WGS method developed by Heitzer et al., using Illumina MiSeq instruments, capable of detecting copy number variations (CNVs) in cfDNA at a depth of 0.1×, with a specificity of over 80% when the ctDNA fraction is ≥10%.
cfDNA Fragmentation-Based Sequencing Methods
This method utilizes whole-genome sequencing to reveal that cancer-derived cfDNA fragments exhibit greater length variability than non-cancer-derived cfDNA. These differences in cfDNA fragment patterns reflect changes in chromatin structure and other genomic and epigenetic abnormalities in cancer.
Multimodal Epigenetic Sequencing Analysis (MESA)
MESA is a flexible and sensitive method that can capture and integrate multiple epigenetic features in cfDNA in a single experiment, such as non-destructive bisulfite-free methylation sequencing.
These methods demonstrate the diversity and importance of cfDNA sequencing in cancer monitoring and management, and with technological advancements, the analysis and application prospects of cfDNA are broad.
References
- Gao Q, Zeng Q, Wang Z et al. Circulating cell-free DNA for cancer early detection. Innovation (Camb). 2022 May 6;3(4):100259. doi: 10.1016/j.xinn.2022.100259. PMID: 35647572; PMCID: PMC9133648.
- Mouliere F. A hitchhiker's guide to cell-free DNA biology. Neurooncol Adv. 2022 Nov 11;4(Suppl 2): ii6-ii14. doi: 10.1093/noajnl/vdac066. PMID: 36380865; PMCID: PMC9650475.
! For research purposes only, not intended for clinical
diagnosis, treatment, or individual health assessments.
 Sources of cfDNA in blood samples. (Florent Mouliere al.,2022)
Sources of cfDNA in blood samples. (Florent Mouliere al.,2022) Future direction for early detection of cancers based on cfDNA tests. (Qiang Gao al.,2022)
Future direction for early detection of cancers based on cfDNA tests. (Qiang Gao al.,2022)


