We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy

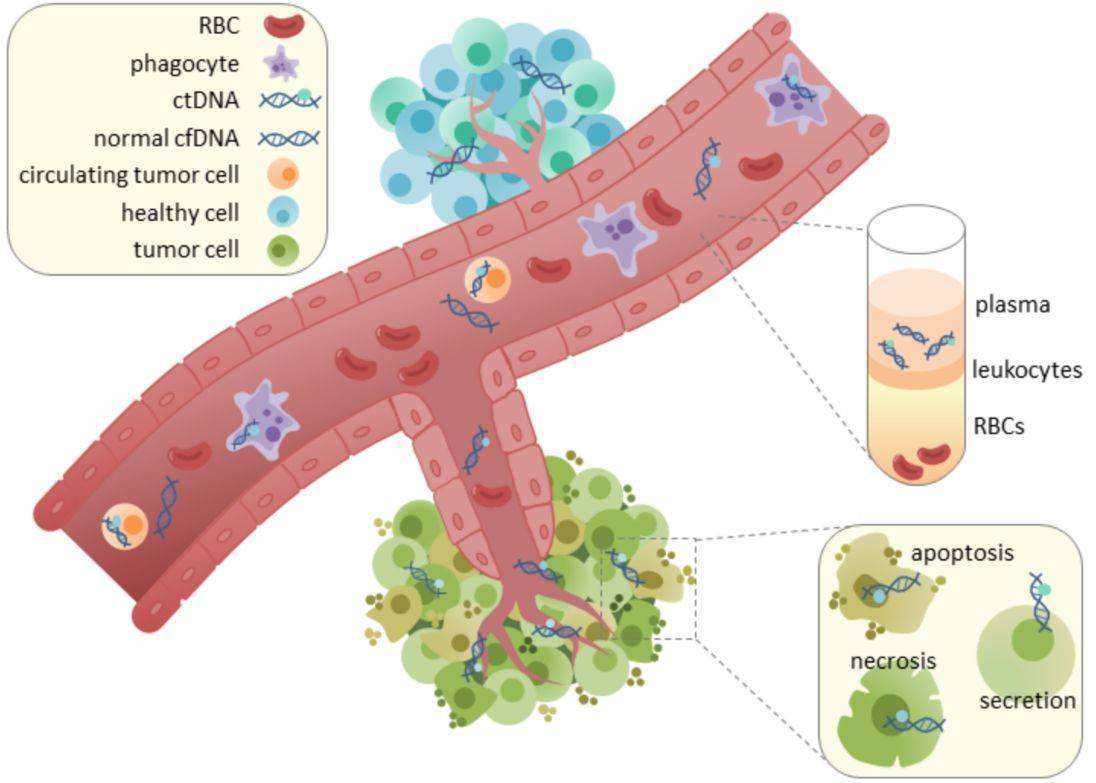
Circulating free DNA (cfDNA), a degraded DNA fragment released by human tissues into the circulatory system such as blood, is a novel molecular marker for tumors.These fragments, which can vary in length between 50 and 300 bp, are generally found at low concentrations in the blood of a healthy individual. The various types of cfDNA isolated from human blood for diagnostic and screening purposes include circulating tumor DNA (ctDNA), cell-free mitochondrial DNA (ccf mtDNA), and cell-free fetal DNA (cffDNA).
ctDNA methylation is one of the important epigenetic modifications that can control gene expression by altering genetic expression without altering gene sequence. Gene methylation is an important mechanism for cancer development, and is a "switch" that regulates gene expression with good stability and consistency.
Changs of level of cfDNA methylation in blood are commonly observed with the progression of cancers and other health conditions. cfDNA analysis has attracted increasing attention because of its easy accessibility, noninvasive nature, and potential tumor specificity through quantitative detection or specific sequencing.
The sensitivity of cfDNA methylation analytic technologies has been greatly improved due to the advancement of molecular biology and next-generation sequencing (NGS) approaches. cfDNA analysis has been widely used in various areas of cancer diagnostics and prognostics, as well as cancer drug resistance, and early screening.
1. Cancer early warning marker detection of precancerous lesions
2. Tumor early screening marker detection
3. Tumor prognosis marker detection
4. Drug efficacy prediction marker detection
CD Genomics has created and verified techniques to perform both specific and complete methylation analysis through sequencing. Additionally, the company has refined the process of separating cfDNA from liquid biopsy specimens to decrease necessary sample amounts for this service.
We can provide cfDNA methylation/ hydroxymethylation detection approaches:
Sample Requirements
|
|
 |
Sequencing Strategy
|
1. Low starting volume
2. High sequencing coverage
3. One-stop services from sample preparation to high-quality data delivery
4. High cost performance
5. Our exclusive ctDNA extraction kit enables precise enrichment of ctDNA fragments
The Standard cfDNA Bisulfite Sequencing Report
- Identification of methylated sites: This report indicates the methylation call and percentage of DNA methylation at each site.
- Statistical analysis results: This report shows the methylated loci with significant differences in methylation levels between groups, along with the annotation of the nearest gene and the relative position of the methylated site to the gene.
- Maps of CpG-rich genomic regions: These maps provide an overview of the distribution of differentially methylated regions across the chromosomes.
- Distribution histograms: This report includes histograms of CpG coverage and CpG methylation levels.
Updated Raw Data
FASTQ files in compressed format
Unique sequences in raw format mapped to the genome (in FASTA file)
Read counts in raw and normalized format, along with the information of mapped reads to each genomic DNA for all queried databases
I hope we've given you some things to think about as you consider using cfDNA in your lab. If you have any requirements do let us know and we will be there to assist you. We'd love the opportunity to work with you. Please contact us for more information and a detailed quote.
References
Terms & Conditions Privacy Policy Copyright © CD Genomics. All rights reserved.