
- Home
- Solutions
- Epigenome Sequencing
- Metagenomics Sequencing
- DRUG-seq Service


B cell receptor (BCR) sequencing is a high - throughput sequencing technique employed to explore the diversity of BCR genes and their functions within the immune system. The BCR, a crucial marker on the surface of B cells, is composed of a heavy chain (IgH) and a light chain (IgK or IgL). Its genes generate highly diverse expression forms through the V(D)J recombination mechanism. BCR sequencing technology has extensive applications in tumor research, immune disease diagnosis, vaccine development, and antibody drug discovery.
The workflow of BCR sequencing primarily involves the amplification and sequencing of genes encoding the BCR in B cells, enabling the analysis of BCR diversity and sequence characteristics. The following is a detailed description of its workflow.
Sample collection and cell separation: Initially, samples containing B cells, such as peripheral blood and bone marrow, are collected. Subsequently, B cells are isolated from other cells using methods like density gradient centrifugation and magnetic bead sorting to obtain a relatively pure B-cell population.
RNA extraction and cDNA synthesis: Total RNA is extracted from the isolated B cells. Using mRNA as a template, reverse transcriptase is employed to reverse - transcribe the RNA into cDNA. This step is essential because the BCR gene is transcribed and expressed in the cell in the form of RNA, which can be converted into DNA through reverse transcription for subsequent amplification and sequencing.
BCR gene amplification: The BCR gene consists of multiple gene fragments, including the variable region (V), diversity region (D), junction region (J), and constant region (C). Specific primers are utilized to perform polymerase chain reaction (PCR) amplification of the BCR gene fragments in the cDNA. These primers are typically designed based on the conserved sequences of the V and J regions, allowing for the amplification of a complete BCR variable - region gene fragment containing the V - D - J junction region. Given the high diversity of BCR genes in the V - D - J junction region among different B cells, PCR amplification can selectively amplify these distinct gene fragments for subsequent detection.
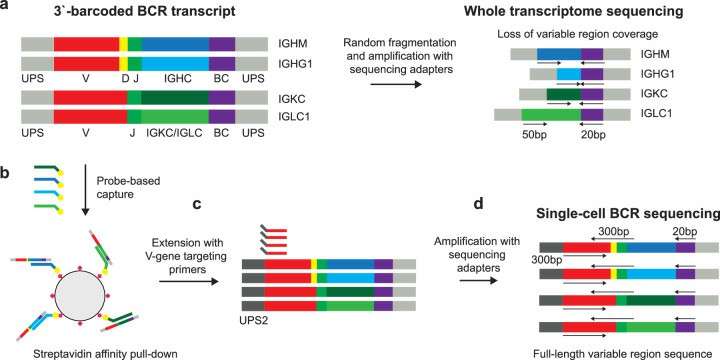 Approach for recovery of full-length variable region BCR sequences from 3'-barcoded sequencing libraries (Morgan et al., 2024)
Approach for recovery of full-length variable region BCR sequences from 3'-barcoded sequencing libraries (Morgan et al., 2024)
Sequencing: The amplified BCR gene fragments are then sequenced. Currently, commonly used sequencing technologies include second - generation sequencing (NGS) and third - generation sequencing. Second - generation sequencing platforms, such as the Illumina sequencing platform, are characterized by high - throughput and low cost. They can sequence a large number of BCR gene fragments simultaneously, generating massive amounts of sequence data. Third - generation sequencing technologies, like PacBio and Nanopore, offer longer read lengths. This feature is particularly advantageous for accurately determining the full-length of the BCR gene, especially in complex gene structures and highly repetitive regions.
Data analysis: The raw data obtained from sequencing undergoes a series of bioinformatics analysis processes. The first step involves quality control of the sequences, where low - quality bases and linker sequences are removed. The processed sequences are then compared with a reference genome to identify the attribution of V, D, and J gene fragments. Additionally, the sequence characteristics of the V - D - J junction region are analyzed, including nucleotide sequence diversity, amino - acid sequence changes, and inserted or deleted bases. Through these analyses, insights into the diversity distribution of B - cell receptors, clone composition, and differences between different samples can be gained. This, in turn, helps to uncover the dynamic changes and specific recognition mechanisms of B cells during the immune response.
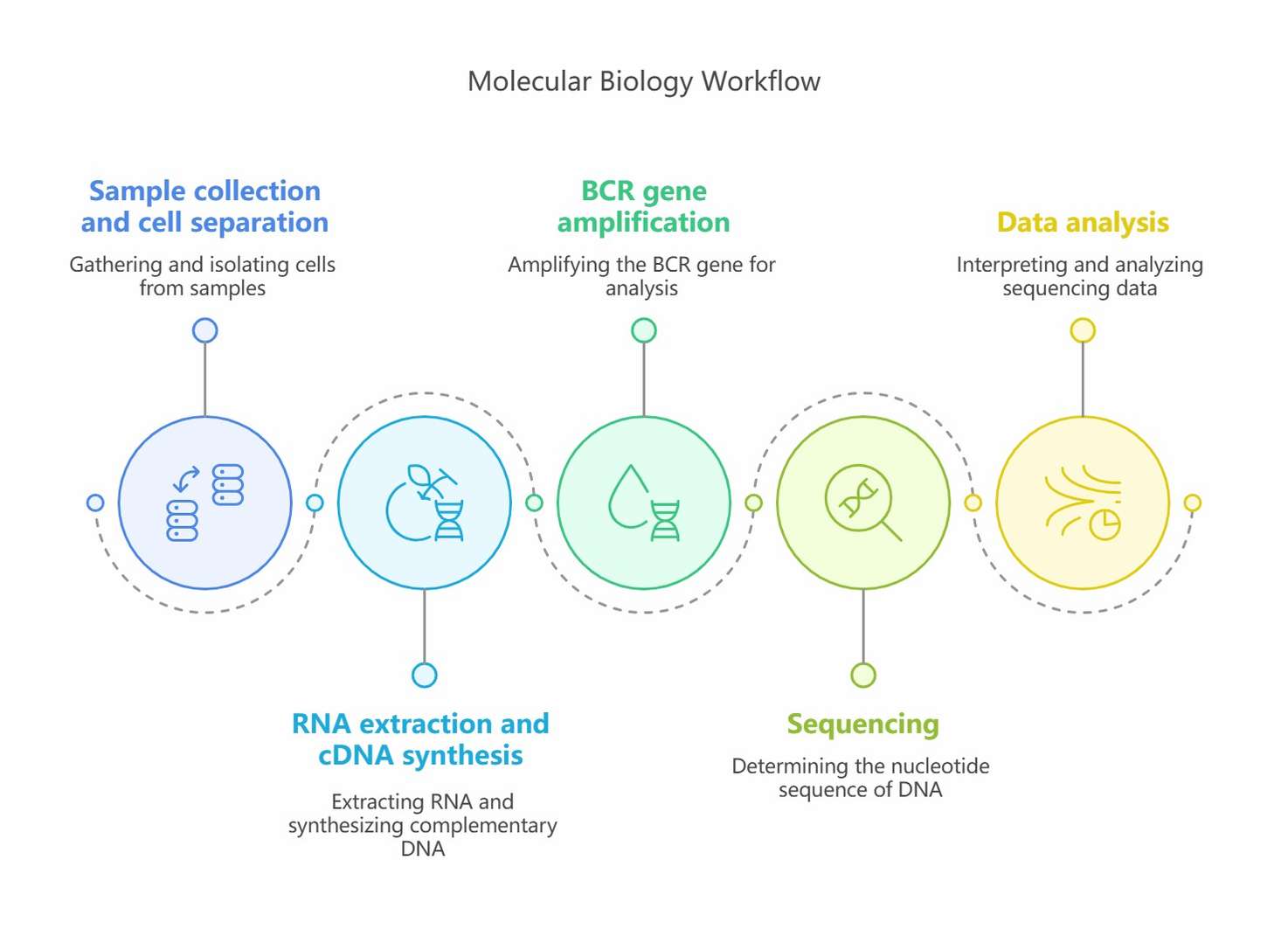 Experimental workflow of repertoire sequencing (Kim et al., 2019)
Experimental workflow of repertoire sequencing (Kim et al., 2019)
Take the Next Step: Explore Related Services
Learn More
From Structure to Function: BCR Classification, Diversity Mechanisms and Their Clinical Implications
BCR sequencing offers several notable advantages. It can detect the diversity of B - cell receptors with high sensitivity and throughput, accurately analyze B - cell clones, and comprehensively and deeply characterize B - cell repertoires in different samples.
High sensitivity and throughput: BCR sequencing technology, via high - throughput sequencing platforms (e.g., Illumina and 10x Genomics), can detect low - abundance clones and mutations. For instance, when detecting rare mutations such as the BCR - ABL fusion gene, next - generation sequencing (NGS) technology can identify more low - frequency mutations in the BCR - ABL kinase region, which are difficult to detect using Sanger sequencing. Moreover, digital PCR (dPCR) also demonstrates higher sensitivity in detecting low - abundance BCR - ABL1 genes.
Comprehensive clonal analysis: BCR sequencing can reveal the clonal origin, diversity, and dynamic changes of B cells. This is of great significance for understanding the immune system's function and disease mechanisms. For example, single - cell sequencing technology enables the tracking of the clonal evolution of a single B cell, uncovering its heterogeneity within the tumor microenvironment. Third - generation sequencing technology further enhances the accuracy of clonal identification, as it can fully cover the full - length BCR sequence, overcoming the limitations of fragment splicing in second - generation sequencing.
Precise disease diagnosis and treatment guidance: BCR sequencing plays a crucial role in disease diagnosis and treatment. In tumor research, it can help identify functional B - cell populations in the tumor microenvironment and uncover the tumor's immune escape mechanism. Additionally, personalized treatment strategies based on BCR sequencing results can significantly improve treatment efficacy.
Integration with other technologies: BCR sequencing can be combined with other technologies to obtain more comprehensive immune system information. When combined with mass spectrometry, it allows for the simultaneous analysis of antibody proteins and BCR repertoires, facilitating a comprehensive understanding of the humoral immune system. Moreover, the combination of BCR sequencing and T - cell receptor (TCR) sequencing can reveal the interaction between T cells and B cells, providing more abundant data for studying the immune response mechanism.
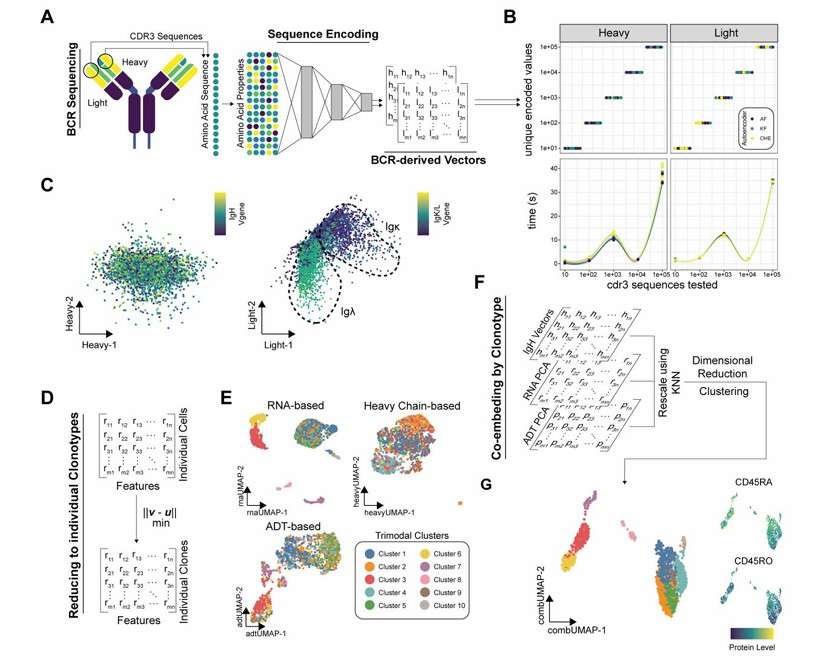 Conceptual schematic for the autoencoding approach for the BCR sequences (Borcherding et al., 2022)
Conceptual schematic for the autoencoding approach for the BCR sequences (Borcherding et al., 2022)
Despite its importance in immune - related disease research, drug development, and other fields, BCR sequencing faces several challenges.
A. Data-related challenges
a) High data complexity: The rearrangement of BCR genes leads to high sequence diversity, accompanied by numerous somatic hypermutations. Additionally, sequencing data contains various noise and errors, increasing the difficulty of data analysis. Specialized bioinformatics tools, such as IgBLAST and Mixcr, can be used to pre - process BCR sequencing data. These tools can remove low - quality sequences, filter noise, and perform gene rearrangement analysis and somatic hypermutation detection, facilitating the extraction of effective information from the data.
b) High data heterogeneity: Differences between different experimental platforms and laboratories pose challenges to data integration and analysis. Adopting cross - center data standardization methods, such as the AIRR - C standard, can standardize data, unifying data formats and quality standards. Technologies like generative adversarial networks (GAN) can also be employed to enhance data balance and reduce the impact of data distribution differences.
B. Experimental challenges
a) Samples acquisition difficulties : In some cases, such as researching rare diseases or B cells in specific tissues, obtaining a sufficient number of high - quality samples can be challenging. For situations where clinical samples are scarce or difficult to obtain, efficient and sensitive database - building technologies, like ImmuHub® 5'RaceUltra technology, can be utilized. This technology enables the successful construction of databases even with a very small number of precious samples. Additionally, the sample size can be increased by optimizing sample collection methods, expanding sample source channels, or collaborating with multiple research institutions for sample sharing.
b) High cost of experimental verification: Verifying the results of BCR sequencing often requires significant time, manpower, and material resources. For example, verifying the function of B - cell receptors through cell culture and protein expression is labor - intensive. Synthetic biological techniques, such as in - vitro TCR sequence synthesis and organ - like co - culture, can be used to simulate the function and immune response of B cells in vitro, reducing verification costs. Moreover, high - throughput verification methods, such as those based on microfluidic chips, can be employed to improve verification efficiency and lower costs.
C. Analytical challenges
a) Difficulty in interpreting biological significance: The biological mechanisms underlying BCR sequencing data are complex. Establishing a connection between sequencing results and the function, immune response, and disease state of B cells to accurately interpret their biological significance is a challenge. Strengthening interdisciplinary cooperation, with biologists, bioinformaticians, and clinicians participating in data analysis and interpretation, is essential. By combining biological experiments, clinical symptoms, and pathological features, a comprehensive analysis of BCR sequencing data can be conducted. Additionally, databases and knowledge graphs can be utilized to integrate existing biological knowledge, assisting in the understanding and interpretation of sequencing results.
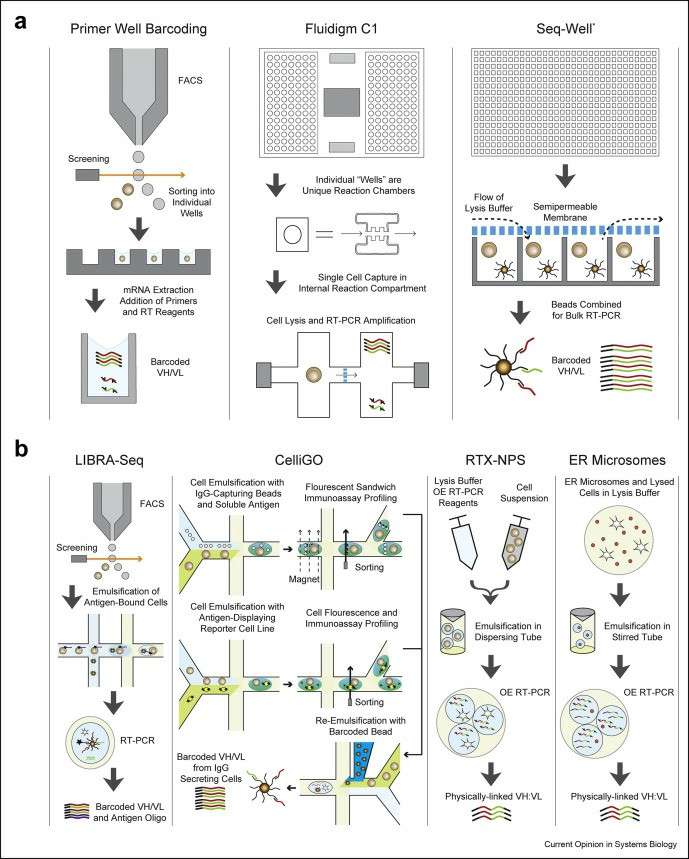 Highlight of methods applicable for Paired BCR sequencing (Curtis et al., 2020)
Highlight of methods applicable for Paired BCR sequencing (Curtis et al., 2020)
BCR sequencing, based on high - throughput sequencing technology, is mainly used to study the diversity of B - cell receptor genes and their functions in the immune system. It has a wide range of applications in tumor research, autoimmune diseases, infectious diseases, vaccine development, and immunotherapy.
Research on B cells development and differentiation
Research on immune response mechanism
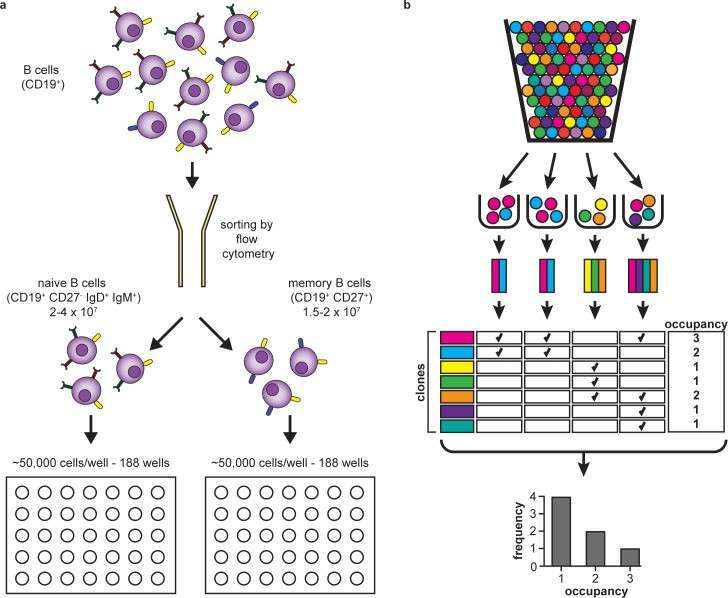 Experimental and informatic design (DeWitt et al., 2016)
Experimental and informatic design (DeWitt et al., 2016)
Disease diagnosis and monitoring
As a vital technology, BCR sequencing has achieved remarkable progress in basic immunology research, disease diagnosis and treatment, and vaccine research and development. Despite its current limitations, with continuous technological advancements and innovations, BCR sequencing is expected to play an even greater role in the future, contributing significantly to human health.
References

CD Genomics is transforming biomedical potential into precision insights through seamless sequencing and advanced bioinformatics.