What is HLA?
Human Leukocyte Antigens (HLAs) are the human version of major histocompatibility complex (MHC). MHC is a set of cell surface proteins essential for the acquired immune system to recognize exogenous molecules in vertebrates, which determines histocompatibility. HLAs are found on all nucleated cells in the body. Each individual has a unique set of HLAs, half derived from each parent.
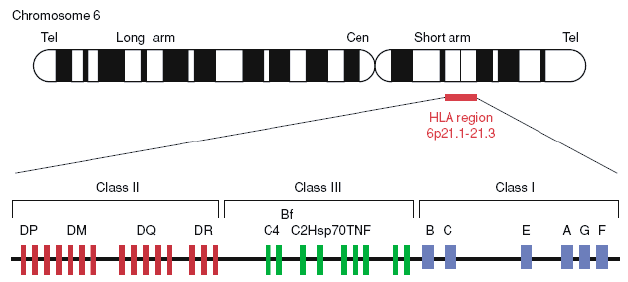 Figure 1. Gene map of the human leukocyte antigen (HLA) region (Berlingerio et al. 2009).
Figure 1. Gene map of the human leukocyte antigen (HLA) region (Berlingerio et al. 2009).
HLA consists of more than 200 genes on chromosome 6 and is categorized into three main groups: HLA class I, HLA class II, and HLA class III. HLA class I genes, including HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, and HLA-G, produce proteins that are bound to peptides on the cell surface of almost all cells. HLA class I can recognize foreign molecules and trigger the infected cell to self-destruct. HLA class II genes, including HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQB1, HLA-DRB1, HLA-DMA, HLA-DMB, HLA-DOA, and HLA-DOB, make proteins that are present almost exclusively on the surface of certain immune system cells. Both MHC class I and class II proteins display peptides to the immune system. However, proteins produced from MHC class III genes function somewhat differently. They are probably involved in immune system activity such as inflammation. And some HLA genes have unknown functions.
The clinical significance of HLA
HLA genes have many variations. For example, HLA-B27 have hundreds of identified alleles. Their high degree of polymorphism allows individual’s immune system to respond to a wide range of foreign invaders, and specific version may be associated with human diseases.
More than 100 diseases have been reported to be connected with different alleles of HLA genes. Many diseases, such as ankylosing spondylitis, liver disease, abnormal immune function, and some types of cancer have been linked with specific HLA alleles.
- HLA system in the clinical treatment
The success of transplantation or transfusion of cells or tissues is strongly associated with the HLA systems of the donor-recipient pair. When donors’ cell surface molecules are different from those present on the recipient, antigens of HLA system elicit strong immune responses since they can be recognized directly or indirectly by the immune system (Figure 2). The direct pathway involves the direct recognition of HLA molecules present on the donor antigen presenting cells (APCs) and the activation of immunologically naive T cells. The indirect pathway involves processing and presentation of donor-derived HLA peptides by the host APC, which operates more efficiently in individuals carrying memory T cells.
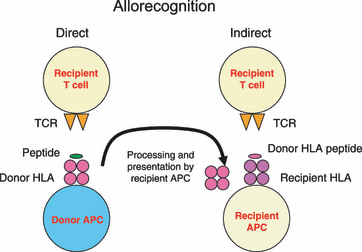 Figure2. Allorecognition (Brown and Navarrete 2011).
Figure2. Allorecognition (Brown and Navarrete 2011).
HLA typing
Since the HLA system has been implicated in the rejection of foreign cells and tissues, HLA typing becomes important in clinical facilities, which allows to predict immune response after transplantation or transfusion. In addition to this, HLA typing is also used as a form of clinical diagnosis to identify markers for specific diseases, such as ankylosing spondylitis. There are two main methods for HLA typing, i.e., detection of HLA antibodies and detection of HLA polymorphism.
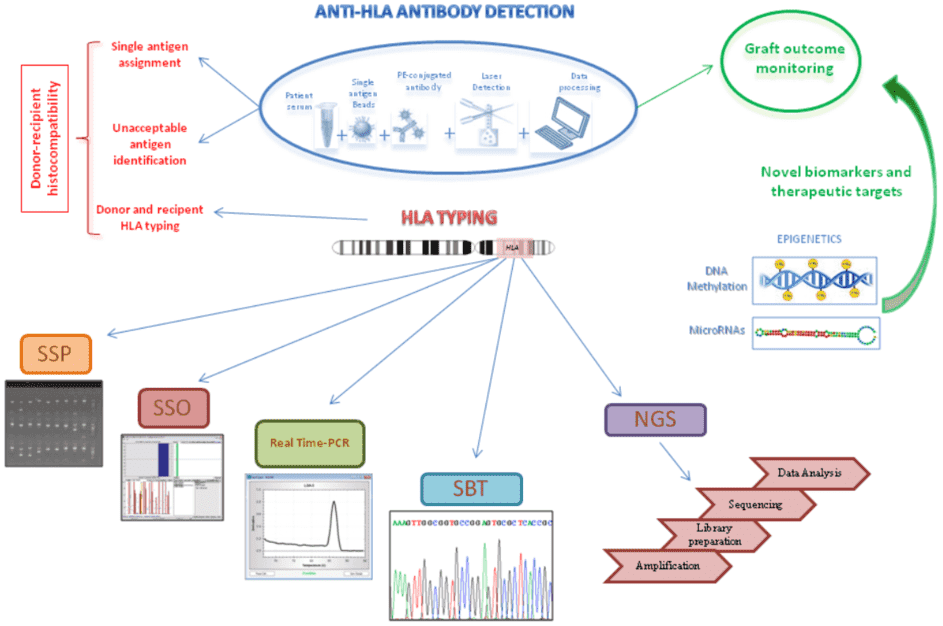 Figure 3. Schematic representation of anti-HLA antibody detection assay by using Luminex technology and the different DNA-based approaches for HLA typing (Picascia et al. 2016).
Figure 3. Schematic representation of anti-HLA antibody detection assay by using Luminex technology and the different DNA-based approaches for HLA typing (Picascia et al. 2016).
- Detection of HLA antibodies
HLA antibodies have been found in about 20% of multiparous women and 30-50% multi-transfused patients. There are three main methods for detection HLA antibodies, including conventional serological cytotoxicity method, flow cytometry, and solid phase technique.
- Serological cytotoxicity method
In this method, magnetic beads are used to isolate B lymphocytes for HLA class II typing from blood or spleen. The remaining leucocytes can be used for HLA class I typing. The lymphocytes are then added to Terasaki plates that hold individual wells containing various specific antibodies. If the specific antibody and HLA antigen bind, the cells in that well will be dead. This method cannot differentiate HLA and non-HLA cytotoxic antibodies.
Here fresh nucleated leucocytes are added to monoclonal antibodies labeled with a fluorescent molecule. Cells with surface HLA antigens that bind to the antibody fluoresces, which can be detected by flow cytometer as they pass through a laser beam.
Fluorochrome-dyed polystyrene beads coated with specific HLA antigens are incubated with the patient’s serum, and the reaction is developed by using a PE-conjugated FC specific antibodies. This method is more sensitive than the serological cytotoxicity method and ELISA, but the clinical benefit of increased sensitivity is not yet clearly defined.
- Detection of HLA polymorphism
With the development of polymerase chain reaction (PCR), DNA-based techniques have dominated in typing laboratories due to their precision and reproducibility as well as enhanced sensitivity and resolution. DNA-based techniques include PCR-sequence specific primers (SSP), PCR-sequence-specific oligonucleotide (SSO), Real-time PCR, Sequencing-Based Typing (SBT), and the next-generation sequencing (NGS).
NGS, also known as high-throughput sequencing, gradually becomes the favorite method for HLA typing for their accuracy, high throughput, and rapidness. The greatest advantage of NGS involves eliminating ambiguities with costs comparable to SBT, and without requiring additional tests. NGS process involves DNA isolation from cells and amplification, DNA library preparation, targeted amplification of genes that code for the HLA peptides using emulsion or solid phase PCR, DNA sequencing-based typing, and sequence alignment against gene bank databases, such as the international ImMunoGeneTics project/human leukocyte antigen (IMGT/HLA) database. High-throughput sequencing-based HLA typing has been widely used in clinical diagnosis, treatment, and personalized medicine.
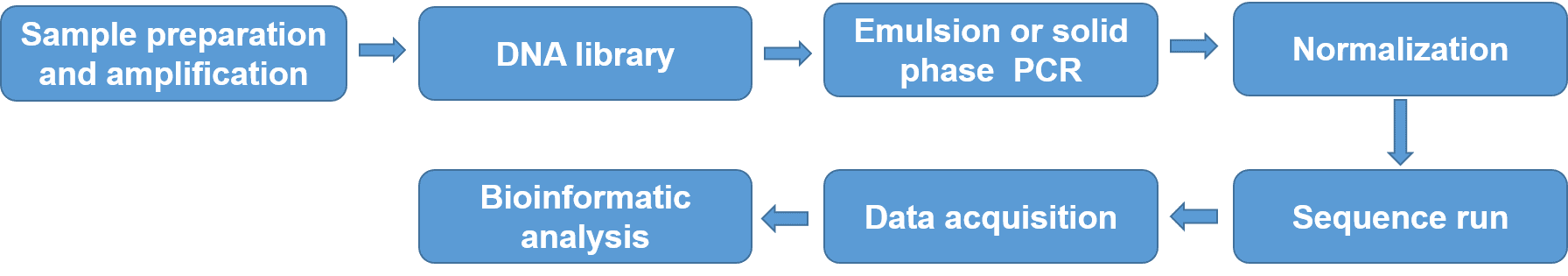 Figure 4. NGS process for HLA typing.
Figure 4. NGS process for HLA typing.
Table 1. NGS HLA-typing software (adapted from 4. Hosomichi et al. 2015).
| HLA-typing software |
URL |
Read type |
Reference |
| HLAminer |
http://www.bcgsc.ca/platform/bioinfo/software/hlaminer |
WGS/WES/RNA-seq/amplicon |
Warren RL et al. |
| seq2HLA |
http://tron-mainz.de/tron-facilities/computational-medicine/seq2hla/ |
RNA-seq |
Boegel S et al. |
| ATHLATES |
https://www.broadinstitute.org/scientific-community/science/projects/viral-genomics/athlates |
WGS/WES/amplicon |
Liu C et al. |
| OptiType |
http://omictools.com/optitype-s6206.html |
WGS/WES/RNA-seq |
Szolek A et al. |
| HLAforest |
http://code.google.com/p/hlaforest |
RNA-seq |
Kim HJ et al. |
| PHLAT |
https://sites.google.com/site/projectphlat/ |
WGS/WES/RNA-seq |
Bai Y et al. |
| Phase-defined HLA sequencing |
https://p-galaxy.ddbj.nig.ac.jp |
Amplicon |
Hosomichi K et al. |
| HLAreporter |
http://paed.hku.hk/genome/software.html |
WGS/WES |
Huang Y et al. |
| HLA-VBSeq |
http://nagasakilab.csml.org/hla/ |
WGS |
Nariai N et al. |
| HLAssign |
http://www.ikmb.uni-kiel.de/resources/download-tools/software/hlassign |
Sequence capture |
Wittig M et al. |
| Assign ATF (Conexio Genomics) |
http://www.conexio-genomics.com |
Amplicon |
— |
| Omixon Target HLA (Omixon) |
http://www.omixon.com/hla/ |
WGS/WES/amplicon |
Major E et al. |
| NGSengine (Gen Dx) |
http://www.gendx.com/products/ngsengine |
Amplicon |
— |
| GeMS (Scisco Genetics) |
http://sciscogenetics.com/services/integrated-genotyping-system/ |
Amplicon |
— |
| HLAscan |
http://www.genomekorea.com/display/tools/HLAscan |
WGS/WES/RNA-seq/amplicon |
Ka S et al. |
At CD Genomics, we are dedicated to providing reliable HLA typing service and professional bioinformatics analysis. Additionally, we also provide services including CRISPR mutation sequencing, antibody screening sequencing, and immune repertoire sequencing, to support drug development and clinical treatment. Please feel free to contact our scientist for detailed service specifications.
References:
- Berlingerio, Michele, et al. "Mining clinical, immunological, and genetic data of solid organ transplantation." Biomedical Data and Applications. Springer, Berlin, Heidelberg, 2009, 211-236.
- Brown C J, Navarrete C V. Clinical relevance of the HLA system in blood transfusion. Vox sanguinis, 2011, 101(2): 93-105.
- Berger. HLA typing. BMJ. 2001 Jan 27:322(7280):218.
- Hosomichi K, Shiina T, Tajima A, et al. The impact of next-generation sequencing technologies on HLA research. Journal of human genetics, 2015, 60(11): 665.
- Ka S, Lee S, Hong J, et al. HLAscan: genotyping of the HLA region using next-generation sequencing data. BMC bioinformatics, 2017, 18(1): 258.
- Picascia, A., Grimaldi, V., Napoli, C. From HLA typing to anti-HLA antibody detection and beyond: The road ahead. Transplantation Reviews, 2016, 30(4), 187-194. doi:10.1016/j.trre.2016.07.007.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1. Gene map of the human leukocyte antigen (HLA) region (Berlingerio et al. 2009).
Figure 1. Gene map of the human leukocyte antigen (HLA) region (Berlingerio et al. 2009). Figure2. Allorecognition (Brown and Navarrete 2011).
Figure2. Allorecognition (Brown and Navarrete 2011). Figure 3. Schematic representation of anti-HLA antibody detection assay by using Luminex technology and the different DNA-based approaches for HLA typing (Picascia et al. 2016).
Figure 3. Schematic representation of anti-HLA antibody detection assay by using Luminex technology and the different DNA-based approaches for HLA typing (Picascia et al. 2016). Figure 4. NGS process for HLA typing.
Figure 4. NGS process for HLA typing.