In the new wave of science and technology, next-generation sequencing (NGS), with its high efficiency and rapidity, shines brilliantly in animal and plant breeding and opens a real genetic revolution. NGS sequencing not only provides Qualcomm quantity and high accuracy DNA sequencing data, but also greatly reduces the cost and time. It is rapidly changing the way of animal and plant breeding and bringing many opportunities for agricultural production.
Introduction of NGS Technologies
Individual genome sequencing: Including Genome Survey, De novo sequencing and telomere-to-telomere (T2T) genome. Using low-depth sequencing data based on small fragment library and K-mer analysis method, Genome Survey can effectively evaluate the important characteristics of a species, such as genome size, GC content, heterozygosity and the proportion of repeated sequences, and can fully understand the genome characteristics of a species, and provide theoretical support for the subsequent de novo sequencing of the whole genome and the formulation of assembly strategies. Genome de novo sequencing refers to sequencing and assembling the genome of a species without relying on the reference genome, so as to draw the whole genome sequence map of the species. T2T genome refers to the assembled telomere-to-telomere genome without gap.
Multi-individual genome sequencing: Including Pan-genome sequencing, bulked segregation analysis sequencing (BSA-Seq), genome-wide association studies (GWAS), Genetic Mapping and population evolution. Pan-genome sequencing focuses on analyzing the genome diversity of all individuals in a species to reveal the genetic diversity of the whole species. Both BSA-seq and GWAS are used to locate genetic markers of specific traits or phenotypes. BSA-seq is mostly used to locate single traits, while GWAS can study multiple traits at the same time. Genetic map is used to determine the physical location of genes on chromosomes, help to understand how genes control specific traits, and reveal genetic variation. The study of population evolution is to use high-throughput sequencing to analyze the genetic variation pattern of species or populations, so as to understand the evolution process and mechanism.
16S, 18S and ITS amplicon sequencing of microbial communities, as well as metagenome sequencing, are the methods to study the whole microbial community in a specific environment. These methods do not need the traditional microbial culture and separation process. By sequencing the DNA of 16S and 18S rRNA genes and amplicon in ITS region or the whole sample, researchers can analyze the microbial community structure, species identification, evolutionary relationship, gene function and metabolic pathway.
RNA sequencing technology includes two main branches: transcriptome sequencing and non-coding RNA sequencing. Transcriptome sequencing focuses on measuring the mRNA expression level of a specific cell or tissue under given conditions, and can also combine the mRNA expression level with SNP for expression quantitative trait loci (EQTL) analysis. Non-coding RNA sequencing involves long non-coding RNA(Lnc RNA), circular RNA(circ RNA), micro RNA (miRNA), small interfering RNA (siRNA), etc. These analyses are helpful to understand the regulation mechanism of genes at epigenetics, transcription and post-transcription levels.
Application of NGS in Plant Research
NGS technology plays a very important role in the study of plants, and it is widely used from basic research to applied research. Its application direction can be briefly summarized as follows:
Basic research applications: Genomics, transcriptomics, epigenetics, system biology and evolutionary biology.
Agricultural applications: Molecular marker-assisted breeding, prediction and selection of genotypes related to excellent traits, plant disease resistance and pathogen detection, and research on diversity and protection of plant germplasm resources.
Ecological and environmental applications: Genetic adaptability of plants in different ecosystems, analysis of response mechanisms under biotic and abiotic pressures, and research on the interaction between plants and microbial communities.
Many technologies of NGS have been applied to improve the breeding of important crops such as wheat, cotton, corn and rice.
Provide foundation for breeding purple testa peanut
Sequencing technology: QTL-seq and BSR-seq
Population construction: 25 strains of Fenpi and 25 strains of Purple Skin F2 were used to construct QTL-seq sequencing pools, 30 purple and 30 green F2 individuals from KF1 population, 10 purple and 10 green F2 individuals from KF2 population and 19 purple and 19 green F7 individuals from ZH population were collected to construct BSR-seq sequencing pool.
Main research contents: Deep purple peanut seed coat is rich in anthocyanins with antioxidant effect, and its nutritional value potential is high. It was found that the purple seed coat of peanut was controlled by a single major gene AhTc1, which encoded a transcription factor R2R3-MYB. Using QTL-seq and BSR-seq techniques, the researchers identified that AhTc1 was located in a 4.7 Mb region on chromosome A10, and developed a specific marker pTesta1089 closely related to purple seed coat. AhTc1 gene is up-regulated in peanut with purple seed coat, and its over-expression in tobacco can lead to purple plant parts. This discovery provides a molecular tool for breeding purple peanut varieties with higher nutritional value.
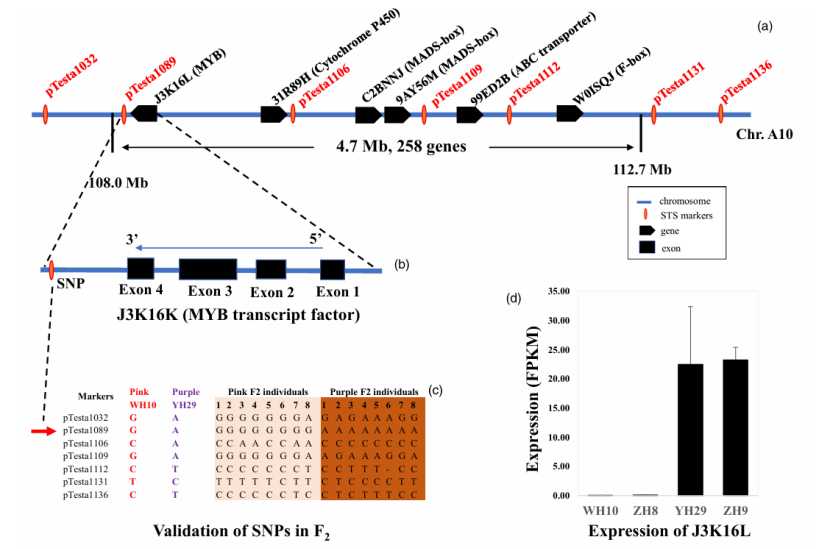 Cloning of AvrStb6 gene from Zymoseptoria tritici (Zhao et al., 2020)
Cloning of AvrStb6 gene from Zymoseptoria tritici (Zhao et al., 2020)
Service you may interested in
Application of NGS in Animal Research
NGS also plays an important role in animal research, mainly focusing on the following aspects:
Basic scientific research: Genome sequencing and assembly, phylogenetic analysis, comparative genomics, genetic diversity research, population genetics and evolutionary dynamics research, etc.
Functional genomics and genetic breeding: Molecular marker-assisted breeding, trait association analysis and genetic resource protection, etc.
Gene expression and regulation research: transcriptome, DNA methylation, histone modification, disease gene identification and drug response.
In recent years, NGS has been widely used in breeding, population evolution, functional gene mining, disease diagnosis and genetic disease screening of pigs, cattle, sheep and poultry.
Bring basis for understanding deleterious mutations
Sequencing technology: Genome-wide sequencing
Research object: 1346 dogs of 161 breeds
Main research contents: Researchers have collected various dog breed data sets to track the genetic markers of dog breed development. By combining genetic distance, migration and genome-wide haplotype sharing analysis, the geographical model of dog breed development and the independent origin of common characteristics were revealed. The study found the mixed history of dog breeds and clarified the influence of immigration on some modern dog breeds. In addition, the study also used phylogeny and haplotype sharing to prove that some common features in the history of dogs appeared more than once. These analyses describe the complexity of the development of dog breeds in detail, and solve the long-term problems about the origin of dog breeds, the migration effect of geographically unique dog breeds, the characteristics of dog breeds and the transfer of disease alleles.
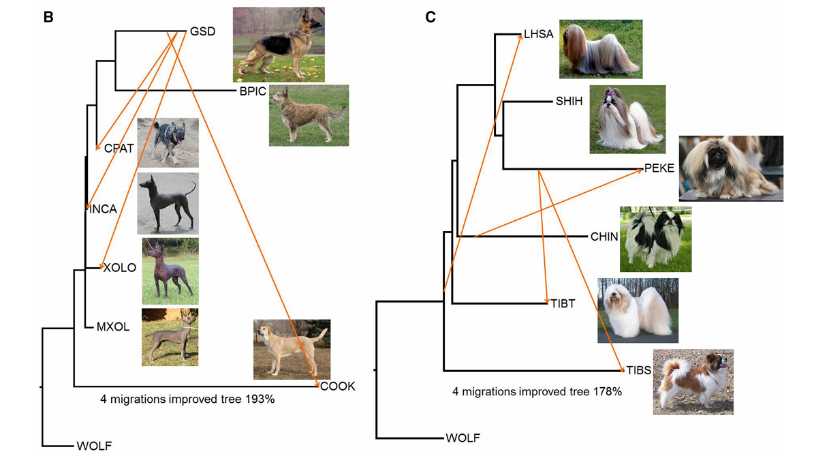 Evolutionary relationship of domestic dogs (Heidi et al., 2017)
Evolutionary relationship of domestic dogs (Heidi et al., 2017)
Prospects of NGS in Agricultural Research
Of course, NGS technology is not limited to genome sequencing (animals, plants and microorganisms), transcriptome sequencing, epigenome sequencing, single cell sequencing and spatial transcriptome sequencing. With the further maturity and price reduction of technology, NGS is expected to be widely used in more scientific research projects in the future. For example, many important traits such as environmental adaptability, stress resistance and nutritional quality can be used for gene mining and functional location through NGS. NGS technology has brought unprecedented opportunities for animal and plant breeding, and provided a tool for researchers to dig and utilize genetic resources in depth.
References
- Zhao Yuhan, Ma Junjie., et al. "Whole-genome resequencing-based QTL-seq identified AhTc1 gene encoding a R2R3-MYB transcription factor controlling peanut purple testa colour." Plant Biotechnology Journal 18 (2020): 96-105. https://doi.org/10.1111/pbi.13175
- Heidi G. Parker, Dayna L. Dreger., et al. "Genomic Analyses Reveal the Influence of Geographic Origin, Migration, and Hybridization on Modern Dog Breed Development." Cell reports 19 (2017): 697-708. DOI: 10.1016/j.celrep.2017.03.079
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Cloning of AvrStb6 gene from Zymoseptoria tritici (Zhao et al., 2020)
Cloning of AvrStb6 gene from Zymoseptoria tritici (Zhao et al., 2020) Evolutionary relationship of domestic dogs (Heidi et al., 2017)
Evolutionary relationship of domestic dogs (Heidi et al., 2017)