CD Genomics offers SNP genotyping using the simple and affordable SNaPshot Multiplex System to allow efficient and quick results.
What is SNaPshot Genotyping
The SNaPshot Multiplex System is a sophisticated SNP genotyping technology developed by Applied Biosystems (ABI). Often referred to as "mini-sequencing," this method is based on fluorescently labeled single-base extension principles. It excels in genotyping multiple single nucleotide polymorphisms (SNPs) simultaneously within a single reaction. SNaPshot's core technology involves using fluorescently tagged dideoxynucleotides (ddNTPs) to terminate the extension of DNA primers after the addition of one nucleotide, allowing for precise determination of SNPs based on the emitted fluorescence.
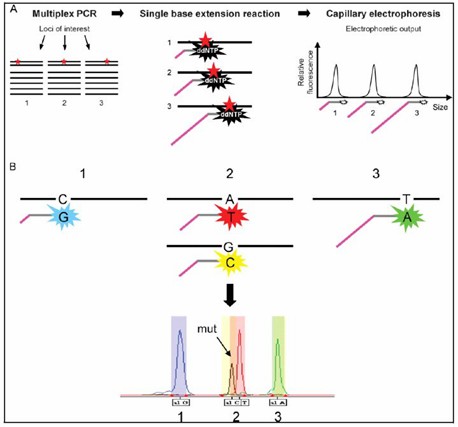 Figure 1. SNaPshot assay principle schematic.
Figure 1. SNaPshot assay principle schematic.
How does a SNaPshot Assay Work
The SNaPshot assay operates through a methodical and efficient workflow that encompasses several crucial steps:
- Primer Design: Specific primers are meticulously designed to anneal immediately upstream of the SNP sites of interest. These primers vary in length, enabling differentiation between SNPs during subsequent analyses.
- Multiplex PCR: In this step, a multiplex polymerase chain reaction (PCR) is performed to amplify regions containing the target SNPs. This involves the use of multiple primers within a single reaction to generate PCR products that encompass multiple SNP loci, thereby enhancing throughput and efficiency.
- Single-Base Extension: Within the reaction mixture, which contains sequencing enzymes and four fluorescently labeled ddNTPs, the primers undergo a single-base extension. Each ddNTP is tagged with a distinct fluorescent dye corresponding to one of the four nucleotides (adenine, thymine, cytosine, guanine) providing a unique signature for each base.
- Detection and Analysis: The extended products undergo capillary electrophoresis, wherein the fluorescent signals emitted by the ddNTPs are detected and recorded. The electropherogram displays peaks where each peak's position correlates to the SNP location and its color identifies the specific nucleotide.
- Data Interpretation: The electropherogram output is analyzed using specialized software to determine SNP genotypes based on the distinct peak colors and positions.
This approach facilitates the simultaneous analysis of up to 16 SNPs in a single reaction, making it highly efficient for medium-throughput genotyping projects.
Introduction of SNaPshot Genotyping Service
CD Genomics offers extensive SNaPshot genotyping services tailored to meet diverse research requirements. Leveraging cutting-edge SNaPshot technology, CD Genomics ensures high-throughput SNP genotyping with unmatched accuracy and efficiency. Our service suite includes:
- Custom Primer Design: Primers specifically tailored to target your SNPs of interest.
- Multiplex PCR and Single-Base Extension: Optimized protocols ensure accurate and reliable outcomes.
- Advanced Capillary Electrophoresis: Use of state-of-the-art technology for precise SNP detection.
- Data Analysis: Expert interpretation of electropherograms utilizing industry-leading software.
Our SNaPshot genotyping service supports projects involving the analysis of 8 to 16 SNPs across several hundred to thousands of samples, making it particularly suitable for large-scale genetic studies and applications.
Advantages of SNaPshot Genotyping Service
SNaPshot genotyping offers several distinct advantages:
- High Accuracy: The SNaPshot method achieves an accuracy rate of up to 99.9%, providing precise SNP genotyping that rivals traditional sequencing methods in reliability.
- Simultaneous Multiplexing: Capable of analyzing up to 16 SNPs in a single reaction, significantly reducing the time and cost associated with genetic analysis.
- Flexibility: Adaptable to a wide range of sample quantities, from a few hundred to several thousand, accommodating varying research scales.
- Broad Applicability: Effective for different SNP types, including those involved in complex genetic traits, without limitations posed by SNP polymorphism characteristics.
- Cost-Effective: Compared to full sequencing approaches, SNaPshot offers an economical solution for high-throughput genotyping, delivering high-quality results at a lower cost.
Applications of SNaPshot Genotyping
SNaPshot genotyping technology finds application in diverse research and clinical contexts, including:
- Genetic Association Studies: Identifying genetic variants associated with diseases or traits.
- Population Genetics: Investigating genetic diversity and evolutionary relationships within and between populations.
- Pharmacogenomics: Exploring genetic factors that influence drug responses, crucial for personalized medicine.
- Agricultural Genetics: Enhancing breeding programs by identifying genetic markers linked to desirable traits in crops and livestock.
The versatility and precision of SNaPshot make it an invaluable tool in both academic research and applied genetic studies, offering robust capabilities for a multitude of genetic investigations.
SNaPshot Genotyping Workflow
Firstly, the primers are used to amplify the target SNPs fragment, and Exo I and the Shrimp Alkaline Phosphatase (SAP) are added to the amplification products to digest the primer and the remaining dNTPs. Then, the purified products are used as templates, and the PCR is performed using the sequencing enzyme, four fluorescent labeling ddNTP and the 5'- terminal extension primers close to the SNP site. The primers extend one base only. After the ABI sequencer, the corresponding SNP loci are determined according to the position and color of the peak. According to the color of the peak, we can know the type of the base, and identify the gene sequence of the sample. It is usually used for analysis of 3 to 30 SNP sites. A typical SNaPshot Multiplex System for SNP genotyping workflow is shown as follow:

Service Specifications
Sample Requirements
|
|
 Click |
Sequencing Strategy
|
| Bioinformatics Analysis We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
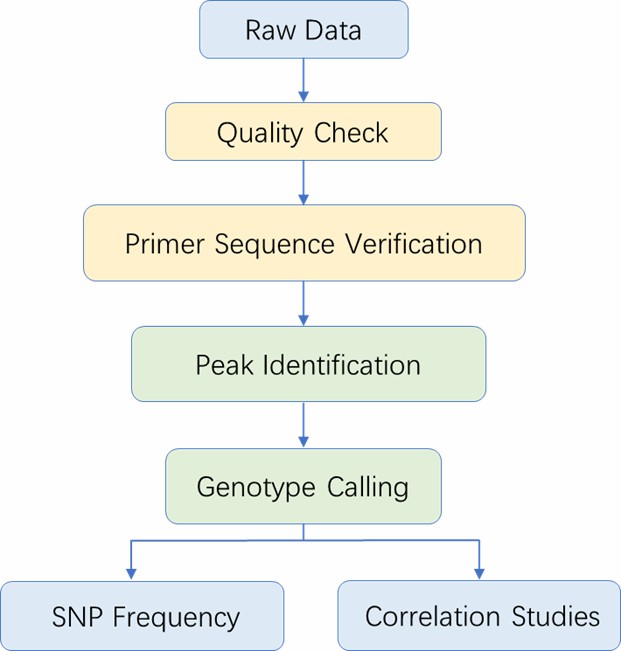
Deliverables
- Raw data files (.fsa)
- Electropherogram files (.pdf)
- Primer sequences used in amplification and reaction systems
- Genotyping results (.excel)
With professional bioinformatics capability, CD Genomics offers high-quality SNP genotyping services for you. If you have any questions, please feel free to contact us.
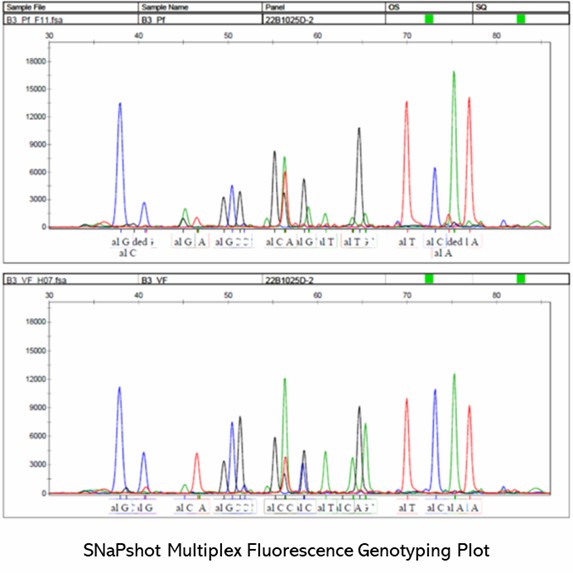
Partial results are shown below:
1. Can SNaPshot detect sample contamination?
Yes, the SNaPshot assay can identify the presence of exogenous DNA contamination by analyzing the height ratios of the dual peaks in the electropherogram. This method provides a reliable means to assess sample integrity.
2. Can the SNaPshot method detect unknown polymorphisms?
Yes, the SNaPshot method is capable of detecting unknown polymorphisms, provided that the sequences flanking the mutation site are known. This allows for accurate base identification at the target site.
3. Can SNaPshot Be Used for Whole-Genome SNP Analysis?
The SNaPshot assay is tailored for targeted SNP genotyping, making it highly effective for analyzing specific SNPs rather than performing a comprehensive whole-genome SNP analysis. For extensive genomic studies that require a broad examination of SNPs across the entire genome, other technologies such as SNP arrays or next-generation sequencing (NGS) offer greater coverage and throughput.
TIMD4 rs6882076 SNP Is Associated with Decreased Levels of Triglycerides and the Risk of Coronary Heart Disease and Ischemic Stroke
Journal: International Journal of Medical Sciences
Impact factor: 3.2
Published: 2019 Jun 2
Background
Coronary heart disease (CHD) and ischemic stroke (IS) are major health concerns linked to atherosclerosis and shared risk factors. This study examined the TIMD4 rs6882076 SNP using Snapshot Technology in 1,765 individuals (581 with CHD, 559 with IS, and 625 controls) to explore its association with CHD, IS, and serum lipid levels.
Materials & Methods
Sample Preparation:
- Human
- Peripheral blood leukocytes
- DNA extraction
Method:
- SNP genotyping
- Snapshot technology platform
- Statistical Analysis
- Allelic frequency
- Chi-square test
- One-way analysis of variance (ANOVA)
Results
The study compared clinical and genetic characteristics between patients with CHD or IS and healthy controls. It found that patients with CHD or IS had higher body weight, BMI, systolic blood pressure, triglycerides (TG), and hypertension rates, but lower total cholesterol (TC), HDL-C, ApoA1, and ApoA1/ApoB ratio compared to controls. The TIMD4 rs6882076 SNP showed different genotypic and allelic frequencies between patients and controls. T allele carriers had a reduced risk of both CHD and IS. In controls, T allele carriers had lower TG levels compared to non-carriers, though this was not significant in patients with CHD or IS. Multivariate analysis identified alcohol consumption, high BMI, and hyperlipidemia as risk factors for both CHD and IS, while smoking was a risk factor for CHD but not IS.
Table 2 Genotypic and Allelic Frequencies of the TIMD4 rs6882076 SNP and the Risk of CHD and IS [n (%)]
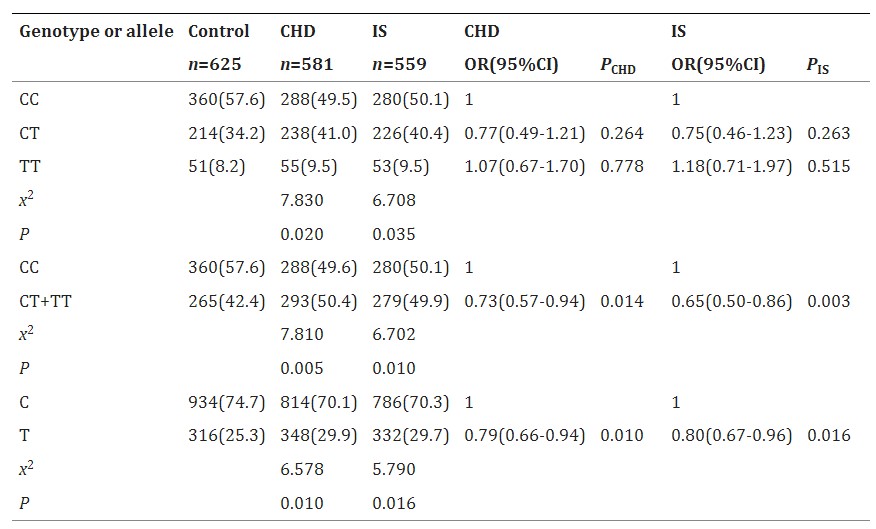
Table 3 The TIMD4 rs6882076 SNP and the Risk of CHD and IS According to Gender, Age, Body Mass Index, Smoking Status and Alcohol Consumption
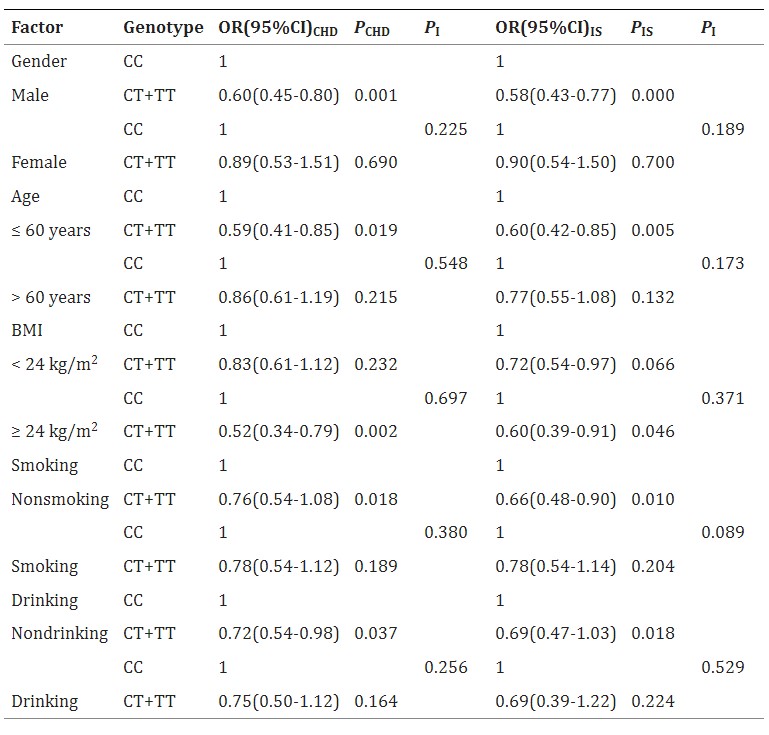
Conclusion
The TIMD4 rs6882076 SNP is linked to lower serum triglyceride (TG) levels and reduced risk of coronary heart disease (CHD) and ischemic stroke (IS) in the Southern Chinese Han population. T allele carriers have a decreased risk of both conditions and lower TG levels compared to non-carriers. This suggests that the SNP may lower CHD and IS risk by reducing serum TG levels.
Reference:
- Khounphinith E, Yin RX, Cao XL, et al. TIMD4 rs6882076 SNP is associated with decreased levels of triglycerides and the risk of coronary heart disease and ischemic stroke. International Journal of Medical Sciences. 2019, 16(6):864.
Here are some publications that have been successfully published using our services or other related services:
Investigating interactive effects of worry and the catechol-o-methyltransferase gene (COMT) on working memory performance
Journal: Cognitive, Affective, & Behavioral Neuroscience
Year: 2021
Eye color prediction using the IrisPlex system: a limited pilot study in the Iraqi population
Journal: Egyptian Journal of Forensic Sciences
Year: 2020
Embryonic origin and genetic basis of cave associated phenotypes in the isopod crustacean Asellus aquaticus
Journal: Scientific Reports
Year: 2023
Scanning indels in the 5q22.1 region and identification of the TMEM232 susceptibility gene that is associated with atopic dermatitis in the Chinese Han population
Journal: Gene
Year: 2017
Impact of the OATP1B1 c.521T>C single nucleotide polymorphism on the pharmacokinetics of exemestane in healthy post-menopausal female volunteers
Journal: Journal of clinical pharmacy and therapeutics
Year: 2017
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
