We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy
Navigation
- Home
- Sequencing
- Genomics Sequencing
- Complete Plasmid/Phage Sequencing
- Mitochondrial DNA (mtDNA) Sequencing
- Viral Genome Sequencing
- Whole Genome Sequencing
- Whole Exome Sequencing
- Targeted Region Sequencing
- Amplicon Sequencing Services
- Chloroplast DNA (cpDNA) Sequencing
- TCR-Seq
- Shallow Whole Genome Sequencing
- Human Mitochondrial DNA (mtDNA) Sequencing
- Long-Read Sequencing
- ctDNA Sequencing Services
- Gene Panel Sequencing Service
- Long Amplicon Analysis (LAA)
- Animal/Plant Exome Sequencing Service
- Transcriptomics
- Bacterial RNA Sequencing
- RNA-Seq
- Small RNA Sequencing
- LncRNA Sequencing
- CircRNA Sequencing
- Ribosome Profiling (Ribo-seq)
- Total RNA Sequencing
- Targeted RNA Sequencing
- Degradome Sequencing
- Exosomal RNA Sequencing
- Ultra Low RNA Sequencing
- Dual RNA-seq
- microRNA Sequencing Service
- mRNA Sequencing Service
- Epigenomics
- Targeted Bisulfite Sequencing
- Reduced Representation Bisulfite Sequencing
- Whole Genome Bisulfite Sequencing (WGBS)
- MeDIP Sequencing (MeDIP-Seq)
- ChIP-Seq
- MeRIP Sequencing (m6A Analysis)
- RIP-Seq
- ATAC-Seq
- NGS-BSP
- DNA 6mA Sequencing
- DAP-Seq Service (DNA affinity purification sequencing)
- 5mC/5hmC Sequencing
- oxBS-seq
- hMeDIP-seq
- Nanopore RNA Methylation Sequencing Service
- RNA Methylation Sequencing Service
- 2'-O-RNA Methylation Sequencing Service
- PacBio SMRT Sequencing
- Microbiome
- Single-Cell Sequencing
- Nanopore Sequencing
- Other Services
- CRISPR Sequencing
- CRISPR Screen Sequencing
- CRISPR Off-Target Validation
- Antibody Screening Sequencing (Phage Display Library Screening)
- Immune Repertoire Sequencing
- Sanger Sequencing
- Pre-made Library Sequencing
- Lentiviral/Retroviral Integration Sites Analysis
- AAV Genome Sequencing
- AAV (Adeno-Associated Virus) Integration Site Analysis
- 10x Spatial Transcriptome Sequencing Service
- HLA Typing
- Genomics Sequencing
- Genotyping
- Population Genetics
- Bioinformatics
- Microarray
- Applications
- Company
- Get Your Instant Quote


 Sample Submission Guidelines
Sample Submission Guidelines
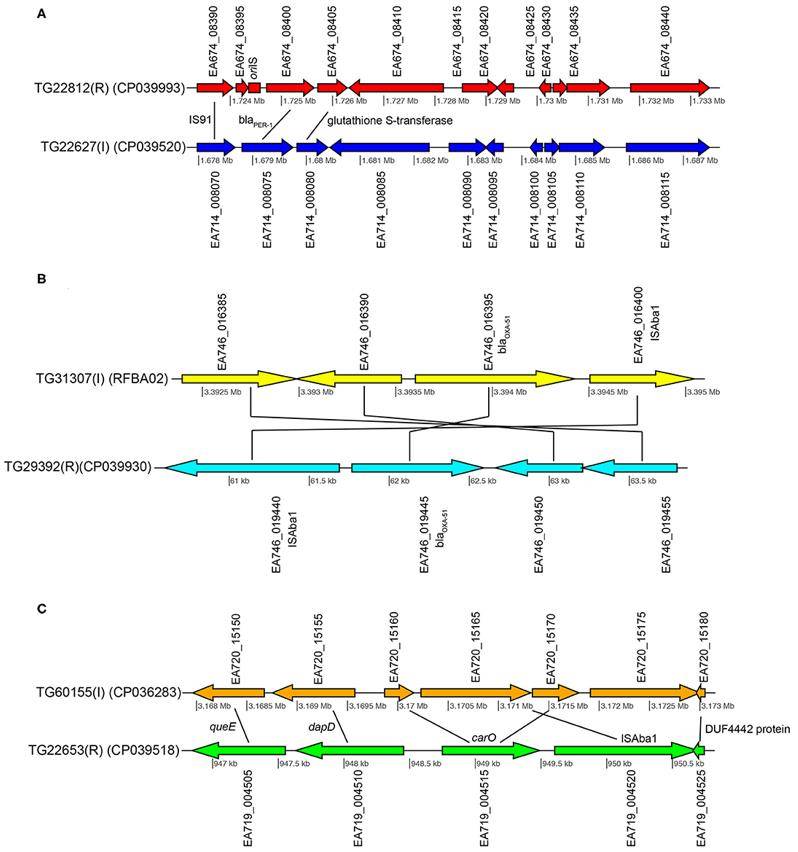 Gene content comparisons between paired isolates (Roe C et al. 2020)
Gene content comparisons between paired isolates (Roe C et al. 2020)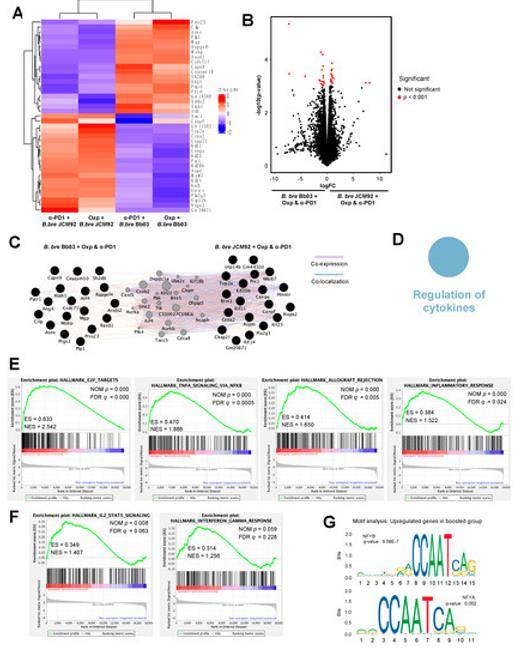 Effect of cancer therapeutics and B. breve strains on the intestinal transcriptome (Yoon Y et al. 2021)
Effect of cancer therapeutics and B. breve strains on the intestinal transcriptome (Yoon Y et al. 2021)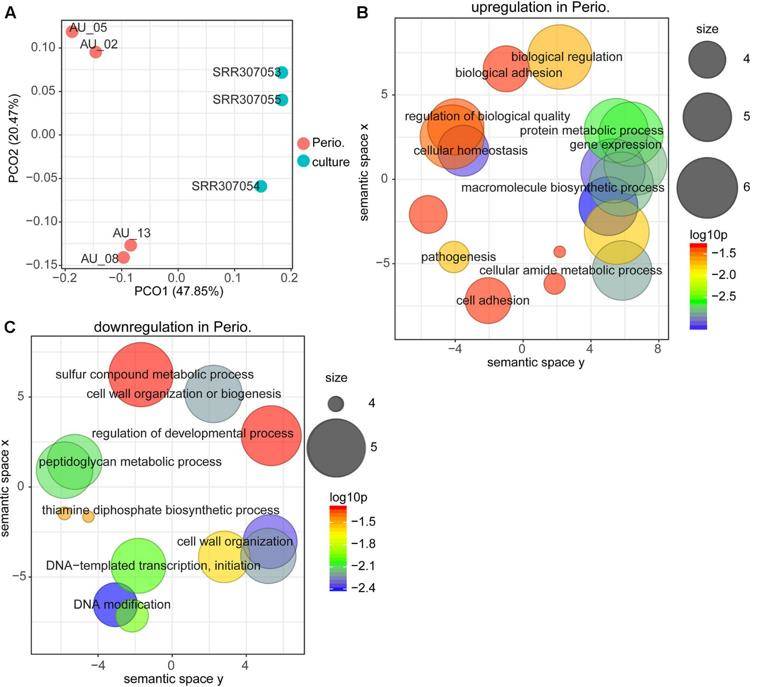 Comparison between gene expression in periodontitis and laboratory culture for Porphyromonas gingivalis (Deng Z L et al. 2018)
Comparison between gene expression in periodontitis and laboratory culture for Porphyromonas gingivalis (Deng Z L et al. 2018)