Introduction
Combining target enrichment with next-generation sequencing (NGS), amplicon sequencing is a rapid and efficient approach that enables researchers to explore genetic variations in specific genomic regions. This method uses specific oligonucleotide probes to target and enrich regions of interest, followed by high throughput sequencing. This highly targeted approach allows ultra-deep sequencing of amplicons (PCR products), enabling accurate and efficient mutation identification and characterization. 16S/18S/ITS amplicon sequencing is widely used for phylogeny and taxonomy studies, particularly in metagenomics samples. Additionally, diverse amplicon panels can be established for disease diagnosis and prognosis.
Advantages
- Enables researchers to discover, validate, and screen variants in a targeted and efficient manner
- Supports multiplexing of thousands of amplicons per reaction to achieve high sequencing coverage
- Allows targeted sequencing even in complex areas, such as GC-rich regions
- Flexible probe designs support a wide range of experiments
- Reduced sequencing costs and turnaround time
Workflow
Figure 1. The workflow of amplicon sequencing.
1. Sample Preparation
Extract DNA from the sample and quantify the DNA.
2. Library Construction
The two-step PCR approach is commonly used to construct amplicon sequencing libraries. In step one, specially designed oligonucleotide probes are used to amplify targeted genomic regions of prepared genomic DNA and attach barcode (to identify amplicons from different samples) to amplicons. In the next step, sequencing adapters are attached. Libraries should be validated and purified to remove additional primers and primer dimers.
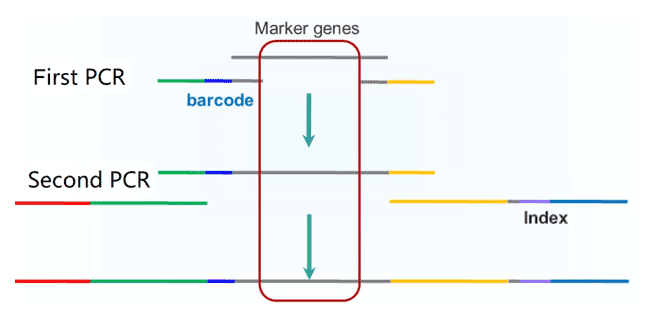
Figure 2. The workflow of two-step PCR for amplicon sequencing.
3. Sequencing
Illumina HiSeq/MiSeq systems and other NGS platforms can be used to sequence amplicons. HiSeq can generate an order of magnitude more reads than MiSeq, but takes longer time.
4. Data Analysis
The quality of the sequenced data needs to be assessed. This step determines which reads should be remained, filtered, trimmed, or removed. High-quality clean data are used for post-analysis.
Table 1. Data analysis of amplicon sequencing.
| Data Analysis |
Details |
| Pre-processing |
Reference genome alignment; data cleanup |
| Variant discovery |
Discovery of SNPs, SNVs, CNVs, and Indels |
| Diversity analysis |
Alpha and beta diversity analysis |
| Taxonomic assignment |
Assign taxonomy to phylogenetically informative marker genes |
| Phylogenetic analysis |
Estimate the evolutionary relationships between detected species |
Applications
Amplicon sequencing offers the flexibility for projects targeting population genetics studies, cancer, and genetic disease, let it be expert-defined or customer-designated. It also enables the discovery of somatic and germline variants.
Amplicon sequencing has been applied to the following fields:
Medical field: discover how human microbes, human health/diseases, and disease-associated genes are related; clinical diagnosis and prognosis; pharmacogenomics; etc.
Agricultural field: explore the interaction between microorganisms and plants or livestock; genetic engineering; marker-assisted breeding; etc.
Environmental field: environmental monitoring and assessment; environmental improvement; etc.
Reference:
- Bybee S M, Bracken-Grissom H, Haynes B D, et al. Targeted amplicon sequencing (TAS): a scalable next-gen approach to multilocus, multitaxa phylogenetics. Genome biology and evolution, 2011, 3: 1312-1323.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
