Background
Rice, a staple food crop that feeds a significant portion of the global population, plays a crucial role in agriculture and food security. Genomic research has been pivotal in enhancing rice breeding and crop improvement. This case study delves into the evolution of the rice reference genome, with a specific focus on the transition from the earlier rice reference genome to the cutting-edge T2T complete reference genome.
The journey of rice genomics began with the release of the initial Nipponbare genome sketch in 2005. This marked the inception of rice research in the genomics era. Subsequently, in 2013, a major milestone was reached with the release of IRGSP-1.0, a reference genome that boasted significantly improved genome assembly results and gene annotation. IRGSP-1.0 quickly emerged as the gold standard reference genome for rice and has been widely utilized in research and breeding endeavors.
Although IRGSP-1.0 was a substantial leap forward, it had its limitations, primarily owing to the constraints of the sequencing technology available at the time. IRGSP-1.0 struggled to assemble complex structural regions, resulting in 72 large gaps (including 19 telomeres), 167 tiny gaps, and 779 unknown bases within the genome. These shortcomings represented approximately 3% of the total genome length, emphasizing the need for further advancements in rice genomics.
Advancements in Genomic Technologies
In recent years, the relentless progress in genome sequencing and assembly technologies has opened new doors for comprehensive genome assembly, including next-generation sequencing, PacBio HiFi, and Oxford Nanopore Technology (ONT) ultra-long read sequencing. While many rice varieties have achieved complete and gapless assembly of their chromosomes, there have still been challenges in addressing 2-5 telomere deletions. Remarkably, except for the maize Mo17 genome, no complex plant or animal genome has achieved a complete, telomere-to-telomere-free assembly of all chromosomes.
The Emergence of T2T-NIP
The key breakthrough in rice genomics came with the development of T2T reference genome of rice, T2T-NIP (version AGIS-1.0). This reference genome was meticulously designed to address the shortcomings of its predecessors and to provide an unprecedented foundation for rice research and breeding. To assess the potential of the rice reference genome (T2T-NIP), this study employed genome resequencing and integrated PacBio HiFi and Oxford Nanopore Technologies (ONT) data from multiple rice populations to conduct population variation analysis, using both AGIS-1.0 and IRGSP-1.0 as reference genomes.
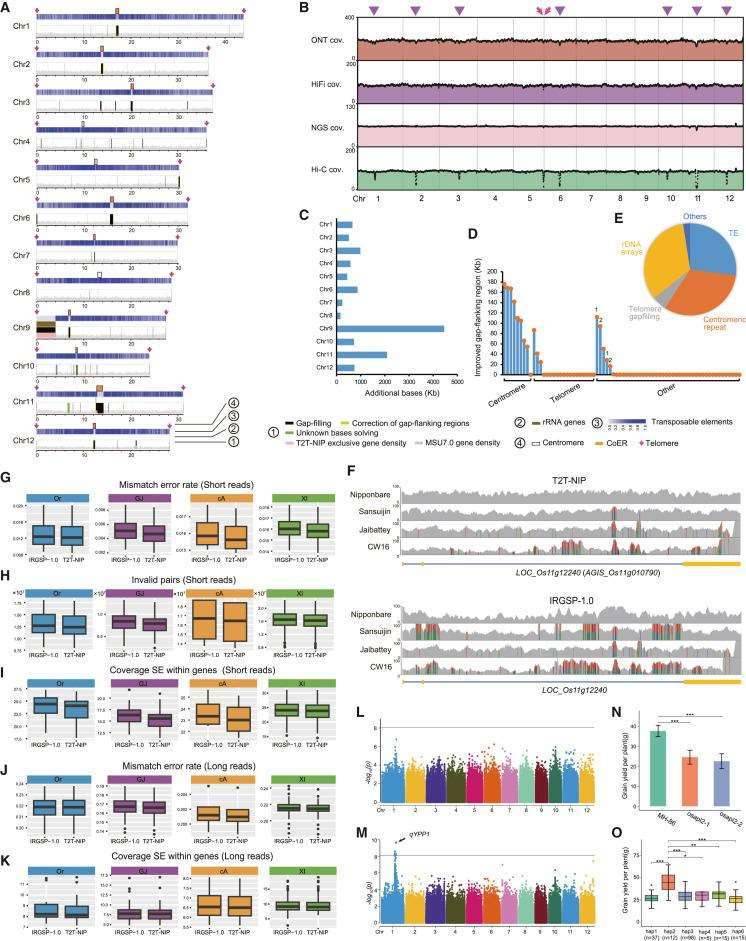 Improvements of the complete rice reference genome T2T-NIP on assembly statistics and potential applications. (Shang et al., 2023)
Improvements of the complete rice reference genome T2T-NIP on assembly statistics and potential applications. (Shang et al., 2023)
The results of this comparative analysis demonstrated the remarkable advantages of AGIS-1.0 over IRGSP-1.0. AGIS-1.0 exhibited higher read utilization rates, improved comparison accuracy, and significantly enhanced comparison quality in regions containing multi-copy genes due to repetitive sequences. Moreover, false-positive comparisons were substantially reduced, enhancing the overall reliability of the reference genome.
Building on the high-quality comparison with AGIS-1.0, the study observed a slight decrease in the number of single nucleotide variants (SNVs) and structural variants (SVs) compared to IRGSP-1.0. Notably, heterozygous variants exhibited a decrease, while purely heterozygous variants showed a consistent increase. These changes were attributed to the improved results of repetitive sequence comparison in AGIS-1.0, further affirming the utility of the new reference genome.
With enhanced accuracy and reduced false positives in variant analysis, the study paved the way for more precise Genome-Wide Association Studies (GWAS). This development in genomics promises to yield valuable insights and accelerate advancements in rice breeding, ultimately contributing to global food security.
The transition from the complete rice reference genome to the AGIS-1.0 reference genome represents a significant milestone in rice genomics. AGIS-1.0 addresses previous limitations and opens up new horizons for research and breeding in the world of rice, offering the promise of improved crop varieties and more sustainable agriculture in the face of global challenges.
Reference:
- Shang, Lianguang, et al. "A complete assembly of the rice Nipponbare reference genome." Molecular Plant 16.8 (2023): 1232-1236.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Improvements of the complete rice reference genome T2T-NIP on assembly statistics and potential applications. (Shang et al., 2023)
Improvements of the complete rice reference genome T2T-NIP on assembly statistics and potential applications. (Shang et al., 2023)