Genomic analysis of cancerous tissues represents one of the most significant developments in modern oncology, providing crucial insights into the molecular basis of malignant diseases. This comprehensive examination delves into tumor genomics as a scientific field, exploring its conceptual foundations and investigative boundaries. Our analysis encompasses the historical progression of genomic research in oncology, cutting-edge methodological approaches, and instrumental technological advances. Furthermore, we investigate how genomic insights translate to clinical practice, particularly in improving diagnostic capabilities and therapeutic interventions. The discussion concludes with an evaluation of current obstacles facing researchers and anticipated developments in tumor genomic studies.
What is Tumor Genomics
Tumor genomics is a scientific discipline dedicated to studying the structure, function, and variations of the tumor cell genome. It plays a central role in cancer research, acting as a precise tool to decipher the genetic information within tumor cells. By meticulously analyzing the tumor genome, researchers can gain insights into genetic alterations occurring during tumor initiation and progression, unveiling crucial molecular clues hidden at the genetic level. These insights not only enhance our understanding of the fundamental nature of tumors but also provide a solid theoretical foundation for developing targeted diagnostic methods, therapeutic strategies, and preventive measures, driving continuous advancements in cancer research.
Research Scope of Tumor Genomics
The research scope of tumor genomics is broad and complex, encompassing DNA, RNA, and protein-level alterations within tumor cells. DNA alterations include gene mutations, chromosomal translocations, and copy number variations, which can activate oncogenes or inactivate tumor suppressor genes, thereby promoting tumorigenesis and progression. At the RNA level, abnormal fluctuations in gene expression influence tumor cell proliferation, apoptosis, and metastatic potential. Proteins, as direct executors of cellular functions, undergo changes in expression levels, post-translational modifications, and interaction networks, playing key roles in tumor progression. These molecular changes are intricately interconnected and collectively determine tumor response to treatment, providing a wealth of therapeutic targets and biomarkers for precision oncology.
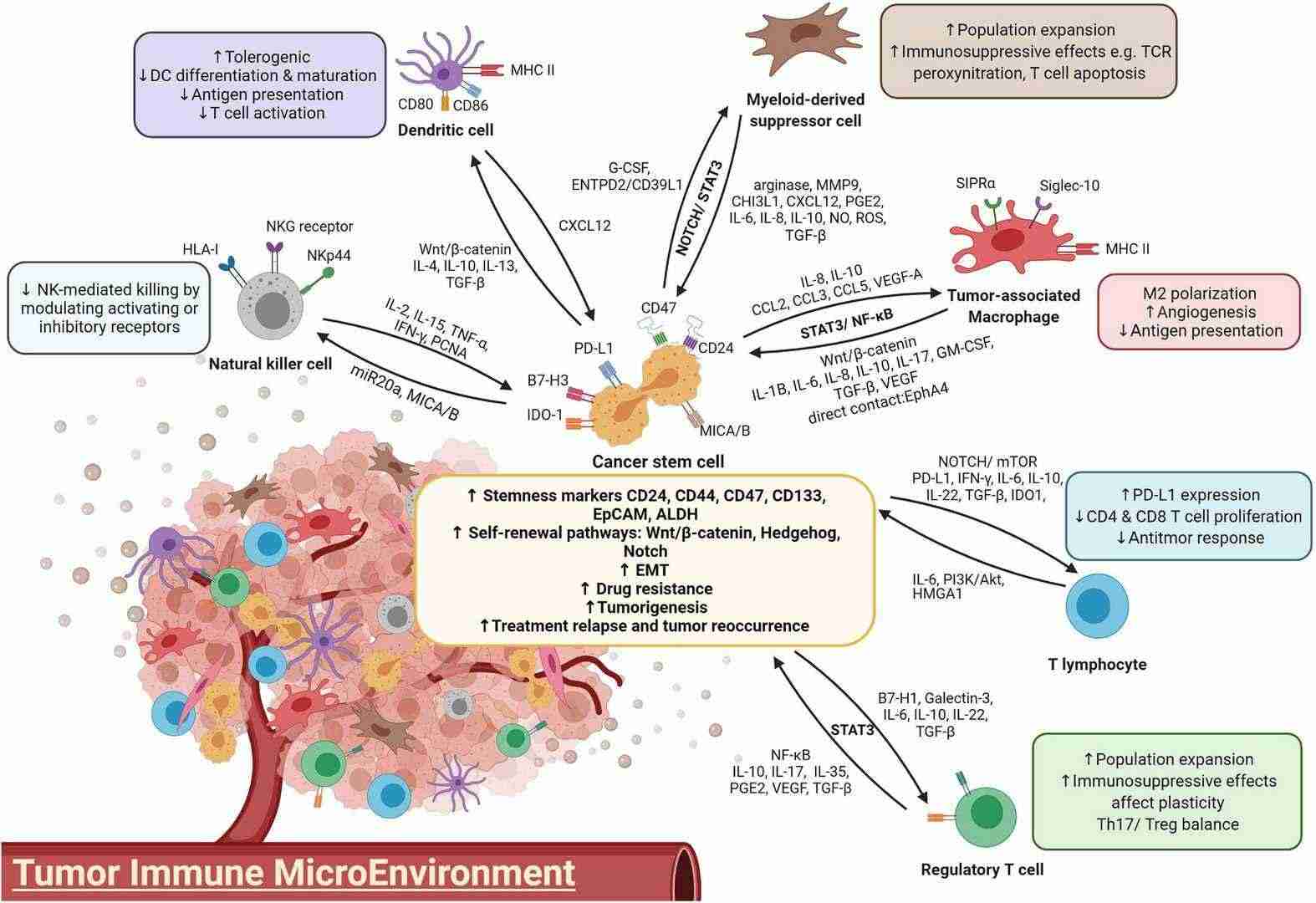 Figure 1. A diagram illustrating the crosstalk between immune cells and CSCs.(Lei, et.al,2021)
Figure 1. A diagram illustrating the crosstalk between immune cells and CSCs.(Lei, et.al,2021)
Service you may interested in
Evolution of Tumor Genomics
The evolution of tumor genomics has witnessed significant research breakthroughs. Initially, scientists observed chromosomal abnormalities in tumor cells through microscopy, laying the foundation for exploring the genetic basis of cancer. With technological advancements, gene cloning and sequencing techniques enabled more precise analysis of cancer-related genes. In the modern era, the advent of high-throughput sequencing technologies has marked a major leap forward. Whole-genome sequencing (WGS) and whole-exome sequencing (WES) have enabled researchers to comprehensively and rapidly identify tumor genome variations. These technological innovations have not only deepened our understanding of cancer mechanisms but have also provided unprecedented opportunities for early diagnosis, personalized therapy, and prognostic assessment.
Research Methods in Tumor Genomics
High-Throughput Sequencing Technologies
High-throughput sequencing technologies play a pivotal role in tumor genomics by providing powerful tools for comprehensive genome-wide analysis. WGS offers an unbiased approach to scanning the entire tumor genome, accurately detecting variations in both coding and non-coding regions, thereby laying the groundwork for an in-depth understanding of tumor genetics. WES focuses on the exonic regions that encode proteins, comprising only about 1% of the genome but harboring numerous cancer-related mutations, enabling rapid identification of key oncogenic mutations. RNA sequencing (RNA-seq) determines RNA sequences and expression levels in tumor cells, reflecting transcriptional activity, revealing tumor-specific gene expression patterns, and identifying novel fusion genes and alternative splicing events. These complementary technologies facilitate a comprehensive understanding of tumor genome variations, providing valuable data for research and clinical applications.
Single-Cell Sequencing Technologies
Single-cell sequencing technologies have revolutionized cancer research by providing a novel perspective on tumor cell heterogeneity and the tumor microenvironment. Single-cell RNA sequencing (scRNA-seq) quantifies gene expression at the single-cell level, unveiling distinct subpopulations within tumors and identifying functionally unique cell subsets, such as cancer stem cells, thereby enhancing our understanding of tumor progression and drug resistance mechanisms. Single-cell ATAC sequencing (scATAC-seq) focuses on chromatin accessibility, elucidating gene regulatory elements and cell-specific transcriptional networks. Together, these techniques offer a detailed view of gene expression and regulatory differences among tumor cells and their interactions with surrounding microenvironmental cells (e.g., immune and stromal cells). This information provides precise targets and theoretical support for developing targeted and immunotherapies, advancing efforts to overcome cancer treatment challenges.
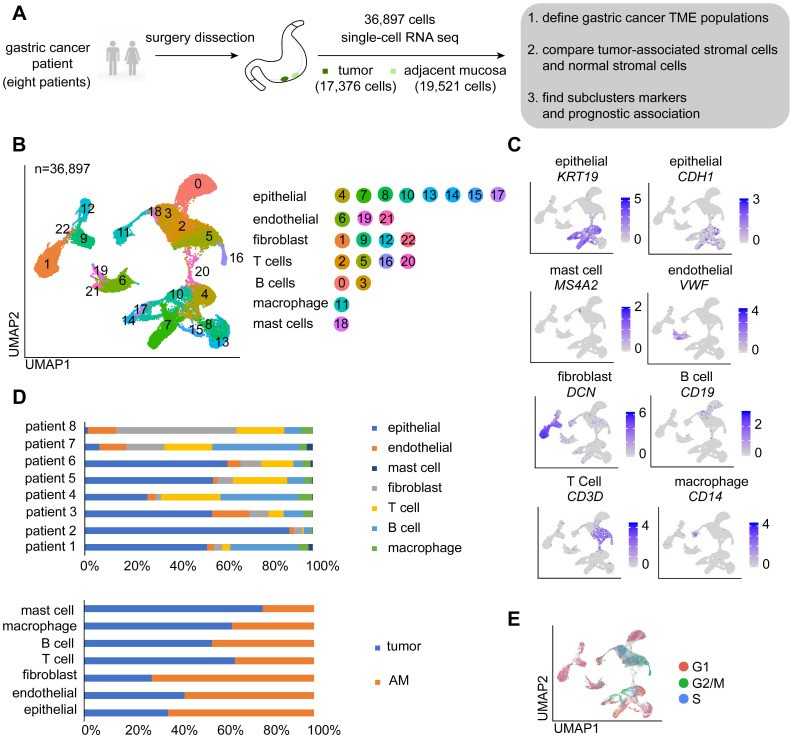 Figure 2. Single-cell sequencing technologies for gastric cancer .(Li, X., et.al,2022)
Figure 2. Single-cell sequencing technologies for gastric cancer .(Li, X., et.al,2022)
Clinical Applications of Tumor Genomics
Early Cancer Detection
Tumor genomics holds immense potential in early cancer detection. Through genomic sequencing, tumor-associated mutations can be systematically scanned, precisely identifying subtle genetic alterations in early-stage tumors. Bioinformatics analysis acts as a sophisticated decoder, extracting valuable information from massive sequencing data to pinpoint specific molecular biomarkers. These biomarkers serve as early warning indicators, signaling tumorigenesis at an incipient stage. For instance, certain mutation signatures frequently appear in early cancer stages, and detecting these mutations significantly enhances diagnostic sensitivity, enabling the identification of tumors at an early and treatable stage. Simultaneously, accurately identified molecular biomarkers improve diagnostic specificity, reducing false-positive rates and granting patients crucial early intervention opportunities, thereby increasing survival prospects.
Prognostic Assessment
Tumor genomics plays a crucial role in prognostic assessment. By analyzing genomic alterations and expression profiles, researchers can construct genetic landscapes that reflect tumor biology. Specific genetic mutations or expression patterns correlate with tumor aggressiveness, metastatic potential, and treatment responsiveness. For instance, the overexpression of certain genes may indicate a higher metastatic risk, while specific mutation combinations may predict sensitivity or resistance to particular therapies. These insights allow clinicians to more accurately predict patient outcomes, stratify patients based on recurrence risk, and tailor follow-up and treatment plans accordingly. This approach enhances disease monitoring, enabling timely interventions and improving long-term cancer management.
Case Studies in Tumor Genomics
Personalized Treatment of Breast Cancer
Tumor genomics plays a critical role in breast cancer treatment, enabling more precise therapeutic strategies for patients. Genetic mutation analysis allows for the identification of key alterations, such as BRCA1 and BRCA2 mutations, which are closely associated with breast cancer development and progression. These genetic insights facilitate the molecular subtyping of breast cancer into categories such as Luminal A, Luminal B, HER2-enriched, and triple-negative breast cancer. Each subtype exhibits distinct biological behaviors and treatment responses. For instance, patients with HER2-enriched breast cancer benefit significantly from targeted therapies such as trastuzumab, which specifically inhibits the HER2 receptor. This targeted approach improves treatment efficacy, prolongs survival, and enhances quality of life, marking a significant advancement in personalized breast cancer treatment.
Esophageal Cancer Research and Treatment
The research team led by Chen Wu has made significant progress in esophageal cancer studies using genomic technologies. Through whole-genome sequencing and transcriptomic analyses, they have identified key genes and signaling pathways involved in esophageal cancer initiation and progression. These discoveries reveal the crucial roles of specific genetic alterations in processes such as cell proliferation, apoptosis, invasion, and metastasis. Furthermore, the team has elucidated the mechanisms of abnormal activation or suppression of these pathways. These findings provide novel molecular biomarkers for early diagnosis, facilitating precise detection of esophageal cancer at its initial stages. Additionally, understanding key genetic alterations and signaling pathways paves the way for developing targeted therapies, offering new treatment options and hope for esophageal cancer patients while advancing therapeutic strategies in this field.
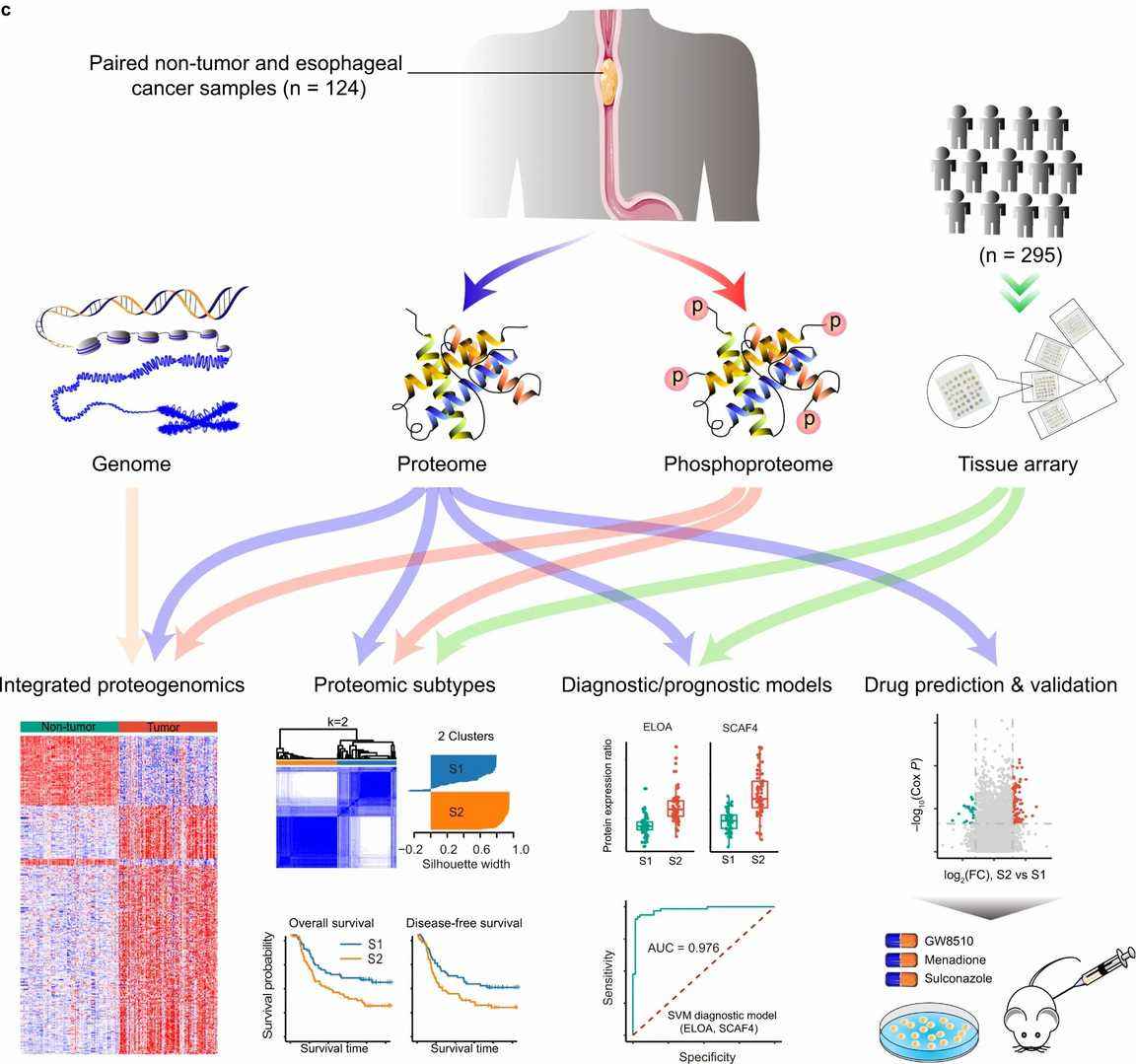 Figure 3. Schematic of the proteomic analyses of esophageal cancer.(Liu, W., et.al,2021)
Figure 3. Schematic of the proteomic analyses of esophageal cancer.(Liu, W., et.al,2021)
Whole-Genome Sequencing in Cancer Research
The research team led by Serena Nik-Zainal at the University of Cambridge has conducted groundbreaking whole-genome sequencing studies on cancer. Utilizing high-throughput whole-genome sequencing technology, they have performed in-depth sequencing on numerous tumor samples from cancer patients. Rigorous data processing and analysis have led to the discovery of novel mutational signatures, significantly enriching the mutational landscape of cancer. These new signatures contribute to more precise classification of cancer types and subtypes, aiding in early diagnosis and providing critical insights for clinicians. Moreover, these discoveries help identify potential targets for novel targeted therapies, driving advancements in personalized cancer treatment. Large-scale whole-genome sequencing projects provide comprehensive and systematic data resources, deepening the understanding of cancer pathogenesis and accelerating innovations in cancer diagnostics and therapeutics.
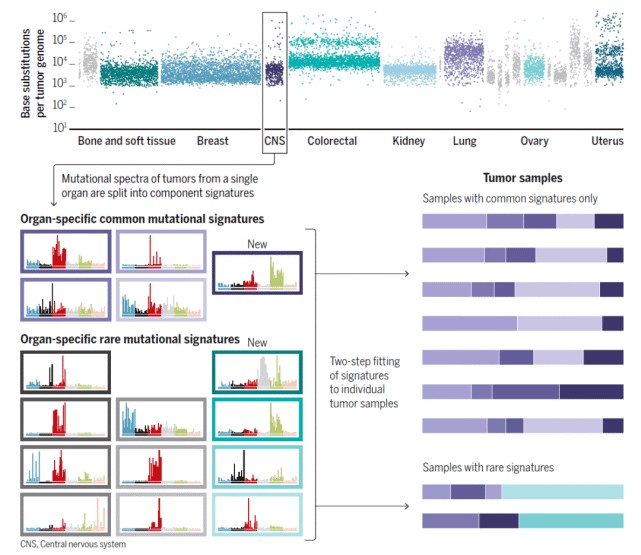 Figure 4. Analysis of characteristic mutations in cancer.(Degasperi, et.al,2022)
Figure 4. Analysis of characteristic mutations in cancer.(Degasperi, et.al,2022)
Future Perspectives in Tumor Genomics
Advances in genomic analysis of tumors present remarkable opportunities for oncological breakthroughs. As technological capabilities expand, enhanced single-cell analysis methods will yield unprecedented insights into the diverse cellular composition within malignancies. By revealing distinct subsets of cells with unique functional properties, these innovations will facilitate more targeted therapeutic approaches in personalized medicine. Examining gene expression through spatial analysis techniques promises to illuminate complex interactions among cells within tumorous environments, deepening our knowledge of structural organization in cancerous tissues and suggesting novel treatment modalities that consider spatial context. Moreover, machine learning applications are becoming central to oncological genomics research. Computational approaches will revolutionize how we process vast genomic datasets, develop predictive frameworks, and generate clinical insights - transforming our ability to detect malignancies early, optimize treatment selection, and forecast patient outcomes. Such technological and analytical innovations are poised to dramatically advance both basic cancer research and clinical practice, ultimately leading to improved patient care through more individualized treatment protocols.
References:
- Lei, M. M. L., & Lee, T. K. W. (2021). Cancer Stem Cells: Emerging Key Players in Immune Evasion of Cancers. Frontiers in cell and developmental biology, 9, 692940. https://doi.org/10.3389/fcell.2021.692940
- Li, X., Sun, Z., Peng, G.,et.al. (2022). Single-cell RNA sequencing reveals a pro-invasive cancer-associated fibroblast subgroup associated with poor clinical outcomes in patients with gastric cancer. Theranostics, 12(2), 620–638. https://doi.org/10.7150/thno.60540
- Liu, W., Xie, L., He, YH. et al. Large-scale and high-resolution mass spectrometry-based proteomics profiling defines molecular subtypes of esophageal cancer for therapeutic targeting. Nat Commun 12, 4961 (2021). https://doi.org/10.1038/s41467-021-25202-5
- Degasperi, A., Zou, X., Amarante, T. D., et.al. (2022). Substitution mutational signatures in whole-genome-sequenced cancers in the UK population. Science (New York, N.Y.), 376(6591), science.abl9283. https://doi.org/10.1126/science.abl9283


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1. A diagram illustrating the crosstalk between immune cells and CSCs.(Lei, et.al,2021)
Figure 1. A diagram illustrating the crosstalk between immune cells and CSCs.(Lei, et.al,2021) Figure 2. Single-cell sequencing technologies for gastric cancer .(Li, X., et.al,2022)
Figure 2. Single-cell sequencing technologies for gastric cancer .(Li, X., et.al,2022) Figure 3. Schematic of the proteomic analyses of esophageal cancer.(Liu, W., et.al,2021)
Figure 3. Schematic of the proteomic analyses of esophageal cancer.(Liu, W., et.al,2021) Figure 4. Analysis of characteristic mutations in cancer.(Degasperi, et.al,2022)
Figure 4. Analysis of characteristic mutations in cancer.(Degasperi, et.al,2022)