Small RNAs (sRNAs), such as microRNAs (miRNAs), piwi-interacting RNAs (piRNAs), and transfer RNA-derived fragments (tRFs), represent a diverse group of non-coding RNA molecules that play pivotal roles in gene regulatory mechanisms. Over the last two decades, advancements in next-generation sequencing (NGS) technologies, particularly RNA-seq and small RNA-seq, have significantly expanded our understanding of small RNA biology. These high-throughput sequencing techniques enable detailed profiling of small RNA expression, making it increasingly essential to study the expression patterns of small RNAs and their differential expression across diverse biological conditions. This review elucidates the role of small RNAs in gene regulation, explicates the scientific rationale for investigating these molecules, outlines the fundamental steps in small RNA differential expression analysis, and examines the bioinformatics tools utilized in these studies. Moreover, it underscores the broad applications of such analyses in both research and clinical contexts, facilitated by the precision and depth provided by NGS methodologies.
The Role of Small RNAs in Gene Regulation
sRNAs play an integral role in the complex regulation of gene expression, acting at both the transcriptional and post-transcriptional levels. Unlike their protein-coding counterparts, sRNAs do not translate into proteins. Instead, they execute their regulatory functions through complex interactions with messenger RNA (mRNA), DNA, and various protein complexes.
miRNAs
Among the diverse classes of small RNAs, miRNAs are particularly prominent, typically consisting of 20-22 nucleotides. These regulatory molecules influence gene expression by binding to complementary sequences on target mRNAs, which leads to either mRNA degradation or the inhibition of translation. MiRNAs are critically involved in a wide array of biological processes, including cellular differentiation, proliferation, and apoptosis. Dysregulation of miRNA expression has been associated with several pathologies, notably cancer, cardiovascular diseases, and neurological disorders.
piRNAs
piRNA mainly works with Piwi proteins in germ cells, playing a key role in silencing transposons and maintaining genome stability. Beyond these roles, piRNAs are implicated in modulating gene expression in response to environmental stressors. Perturbations in piRNA expression are correlated with reproductive and developmental disorders, underscoring their critical functional significance.
tRFs
tRFs, a category that includes tRFs and tRNA-derived stress-induced RNAs (tiRNAs), have gained recognition as vital modulators of gene expression. Arising from precursor tRNAs, especially under stress conditions, these fragments influence translation and cellular stress responses, with emerging evidence to tumorigenesis. The study of tRFs in gene regulation is growing quickly, showing the complexity of small RNA regulatory networks.
Small Nucleolar RNAs (snoRNAs) and Small Nuclear RNAs (snRNAs)
Traditionally known for their roles in ribosome biogenesis and RNA processing, small nucleolar RNAs (snoRNAs) and small nuclear RNAs (snRNAs) are now being acknowledged for their broader regulatory effects on gene expression. Recent studies suggest that their functions extend significantly beyond conventional roles in rRNA modification and pre-mRNA splicing, indicating a versatile and impactful presence in the regulatory landscape.
CDG specializes in next-generation sequencing and transcriptomics, providing advanced small RNA differential expression analysis and customizable bioinformatics solutions to support your research needs.
Our services include:
Rationale for Investigating Small RNA Expression
Small RNA (sRNA) regulation of gene expression is a complex and dynamic process that is essential for maintaining cellular balance and supporting proper organism development. Given their widespread involvement in a myriad of biological functions, studying small RNA expression helps us understand the underlying mechanisms of gene regulation."
Disease Diagnosis and Therapeutic Applications
Aberrant expression patterns of small RNAs, especially miRNAs, have been associated with a diverse array of pathological conditions. MiRNAs have emerged as both potential biomarkers for disease diagnosis and as viable targets for therapeutic intervention. For instance, the dysregulated expression of particular miRNAs has been documented in numerous cancer types, rendering them prospective targets for innovative therapeutic strategies. Additionally, the development of small RNA-based therapies, including miRNA mimics or inhibitors, is currently under investigation in clinical trials for diseases such as cancer, viral infections, and autoimmune disorders.
Stress Response and Adaptation to Environmental Changes
Small RNAs, notably piRNAs and tRFs, play significant roles in modulating cellular stress responses and adaptation to environmental perturbations. The expression levels of these small RNAs may vary in response to stressors such as thermal shock, hypoxic conditions, or oxidative stress. A comprehensive understanding of these expression changes can elucidate how cells and organisms adapt to environmental stressors, presenting potential applications in agricultural biotechnology, environmental surveillance, and the prevention of stress-related diseases.
Influence on Gene Regulatory Networks
Small RNAs are crucial constituents in the formation and maintenance of gene regulatory networks. By influencing the expression of transcription factors and other pivotal regulators, small RNAs can modulate entire signaling pathways. Investigating the differential expression of small RNAs across varied biological contexts allows researchers to pinpoint critical regulatory nodes within these networks, furnishing invaluable targets for drug development and advancing functional genomics research.
Key Steps in Differential Expression Analysis of Small RNAs
Analyzing small RNA expression and identifying differentially expressed small RNAs are intricate tasks necessitating meticulous experimental design, data curation, and statistical analysis. Below are the key steps involved:
1. Data Preparation and Quality Control
Prior to commencing differential expression analysis, the small RNA sequencing data must be prepared meticulously, entailing:
- Raw Data Acquisition: Small RNA sequencing data is typically procured through high-throughput sequencing technologies such as Illumina sequencing, producing millions of short reads that represent small RNA molecules in the sample.
- Preprocessing: The raw sequencing reads are processed to eliminate low-quality reads, adapter sequences, and any contaminant sequences, ensuring a high-quality dataset for downstream analysis.
- Normalization: To accommodate variations in sequencing depth across samples, data normalization is imperative. This step is critical to ensure that observed differences in small RNA expression reflected biological variance rather than technical discrepancies.
2. Expression Matrix Construction
Once data is cleansed and normalized, the next step involves constructing an expression matrix, representing the counts of each small RNA in each sample. Bioinformatics tools like DESeq2 are typically employed to convert raw read counts into a normalized expression matrix, primed for differential expression analysis.
3. Statistical Analysis
The core of differential expression analysis entails comparing small RNA expression levels between experimental groups. Tools like DESeq2 or edgeR are predominantly used, modeling count data statistically, such as by employing the negative binomial distribution, to perform hypothesis testing and identify differentially expressed small RNAs.
- DESeq2: This tool applies a negative binomial distribution to model count data, employing empirical Bayes methods to shrink log fold changes, enhancing differential expression analysis accuracy, particularly with small sample sizes. DESeq2 also facilitates result visualization using tools like volcano plots and heatmaps for interpreting differential expression patterns.
- edgeR: Widely employed for RNA-seq data analysis, edgeR utilizes a similar statistical framework as DESeq2 but is often favored for analyses involving fewer biological replicates or smaller RNA datasets.
4. Result Visualization and Interpretation
Post-analysis, result visualization is crucial for interpretation. Common visualization tools include:
- Volcano Plots: Offering a visual summary of differential expression outcomes, these plots highlight the most significant expression changes based on p-value and fold change.
- Heatmaps: Used to visualize small RNA expression levels across different samples, heatmaps provide insights into gene expression patterns and facilitate clustering of samples with analogous expression profiles.
By carefully analyzing small RNA expression, we can uncover the complexities of gene regulation, which has important implications for research and clinical applications.
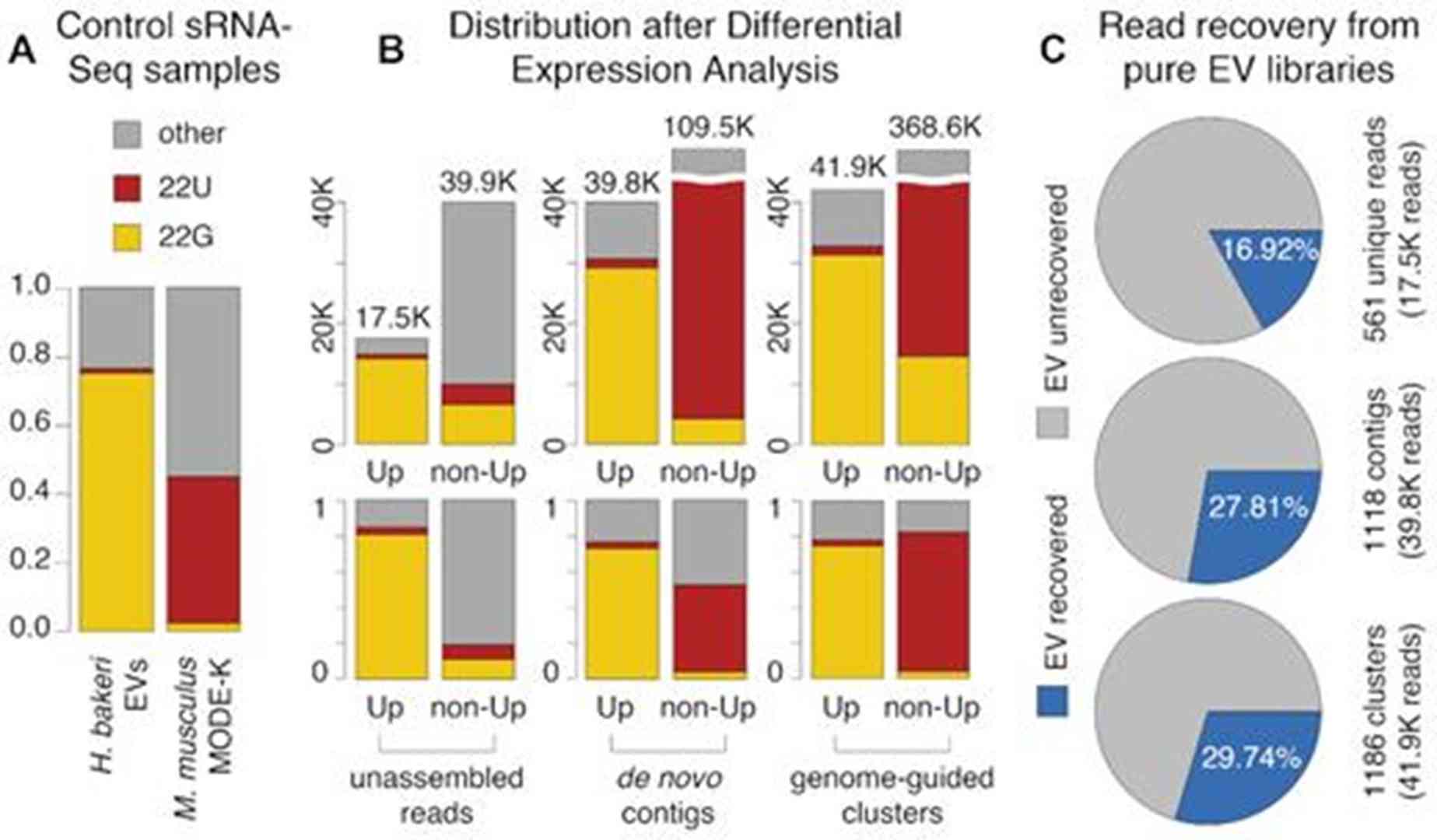 Evaluation of differential expression results. (Bermúdez-Barrientos, et al. Nucleic Acids Research, 2020)
Evaluation of differential expression results. (Bermúdez-Barrientos, et al. Nucleic Acids Research, 2020)
Bioinformatics Instruments for Small RNA Analysis
The landscape of small RNA analysis is enriched by an array of bioinformatics tools, each tailored with distinct capabilities to address various research inquiries.
DESeq2
Among the stalwarts of RNA sequencing differential analysis, DESeq2 is renowned for its capability to manage high-dimensional count data intrinsic to small RNA sequencing. It incorporates robust statistical frameworks to discern differentially expressed small RNAs across diverse conditions. Furthermore, DESeq2 seamlessly integrates with visualization tools such as pheatmap and ggplot2, facilitating the interpretation of analytical outcomes.
edgeR
Esteemed for its efficacy in handling datasets with limited sample sizes, edgeR stands out as a preferred choice for RNA-seq analysis. Sharing with DESeq2 the utilization of a negative binomial distribution model, edgeR excels in hypothesis testing to pinpoint significant variances in expression levels.
miRDeep2
miRDeep2 specializes in the discovery of novel miRNAs from high-throughput sequencing datasets. Its utility is particularly pronounced in exploring small RNA expression within organisms or experimental conditions where miRNA datasets remain scantily characterized.
Small RNA Workbench
This comprehensive toolset caters to the end-to-end analysis of small RNA sequencing, encompassing quality control, sequence alignment, and differential expression analysis. It serves as an optimal choice for researchers seeking a cohesive workflow tailored to small RNA analysis.
iDEP (Integrated Differential Expression and Pathway Analysis)
iDEP distinguishes itself as an accessible web application incorporating R/Bioconductor packages with extensive annotation databases for over 200 species. It simplifies RNA-Seq data analysis, offering functionalities for both differential expression and pathway analysis. Notably, iDEP supports reproducible research workflows, empowering biologists to derive molecular insights with minimal reliance on bioinformatics expertise, aiding in the formulation of new hypotheses and validating established pathways.
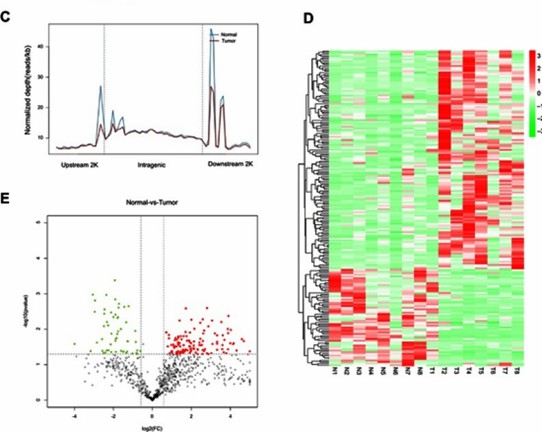 (D) Heatmap depicting the differential methylation of miRNA-encoding genes. (E) Volcano plot illustrating the differential expression of reads between tumor and normal tissues. (Gao, X et al., OncoTargets and Therapy, 2019)
(D) Heatmap depicting the differential methylation of miRNA-encoding genes. (E) Volcano plot illustrating the differential expression of reads between tumor and normal tissues. (Gao, X et al., OncoTargets and Therapy, 2019)
Applications of Small RNA Differential Expression Analysis
The differential expression analysis of small RNAs spans a broad spectrum of applications, impacting both fundamental research and clinical practice.
Cancer Research
The profiling of small RNA expression is increasingly pivotal in identifying biomarkers crucial for cancer diagnosis, prognosis, and therapeutic interventions. Variations in miRNA expression correlate with multiple cancer types, revealing pathways for developing diagnostic assays and innovative therapies.
Neurological Disorders
Dysregulation in small RNA expression is associated with neurological conditions, including Alzheimer's, Parkinson's, and neurodevelopmental disorders. Profiling differentially expressed small RNAs in affected individuals explains molecular mechanisms underpinning these diseases and aids in identifying viable therapeutic targets.
Responses to Stress and Disease
Small RNAs play an essential role in orchestrating responses to environmental stressors such as temperature fluctuations, hypoxia, and oxidative challenges. Analyzing differential small RNA expression in stress contexts yields insights into cellular adaptation, with broader implications for enhancing stress resilience in plants, understanding disease resistance mechanisms, and innovating treatments for stress-associated ailments.
Agriculture and Crop Enhancement
Small RNA regulation is integral to plant growth dynamics, developmental processes, and responses to biotic and abiotic stressors. Profiling small RNA expression in plants can unveil genes linked to drought tolerance, disease resistance, and yield enhancement, thereby supporting progressive strides in agricultural biotechnology.
Conclusion
Small RNA differential expression analysis emerges as a powerful investigative medium for unraveling the intricate regulatory networks governing gene expression. Augmented by cutting-edge bioinformatics tools and high-throughput sequencing methodologies, the exploration of small RNA dynamics has yielded profound insights into diverse biological phenomena. As the inquiry into small RNAs' functional roles advances, their application in disease diagnostics, therapeutic innovations, and agricultural biotechnology promises expansive potential for groundbreaking discovery and innovation.
References:
- Spies, Daniel, et al. "Comparative analysis of differential gene expression tools for RNA sequencing time course data." Briefings in bioinformatics 20.1 (2019): 288-298. https://doi.org/10.1093/bib/bbx115
- Quinn, Thomas P., Tamsyn M. Crowley, and Mark F. Richardson. "Benchmarking differential expression analysis tools for RNA-Seq: normalization-based vs. log-ratio transformation-based methods." BMC bioinformatics 19 (2018): 1-15. https://doi.org/10.1186/s12859-018-2261-8
- McCormick, K.P., Willmann, M.R. & Meyers, B.C. Experimental design, preprocessing, normalization and differential expression analysis of small RNA sequencing experiments. Silence 2, 2 (2011). https://doi.org/10.1186/1758-907X-2-2
- Ge, S.X., Son, E.W. & Yao, R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics 19, 534 (2018). https://doi.org/10.1186/s12859-018-2486-6
- Gao, Xin, et al. "Exploration of bladder cancer-associated methylated miRNAs by methylated DNA immunoprecipitation sequencing." OncoTargets and therapy (2019): 6165-6174. https://doi.org/10.2147/OTT.S192248
- Bermúdez-Barrientos, José Roberto, et al. "Disentangling sRNA-Seq data to study RNA communication between species." Nucleic Acids Research 48.4 (2020): e21-e21. https://doi.org/10.1093/nar/gkz1198


 Sample Submission Guidelines
Sample Submission Guidelines
 Evaluation of differential expression results. (Bermúdez-Barrientos, et al. Nucleic Acids Research, 2020)
Evaluation of differential expression results. (Bermúdez-Barrientos, et al. Nucleic Acids Research, 2020) (D) Heatmap depicting the differential methylation of miRNA-encoding genes. (E) Volcano plot illustrating the differential expression of reads between tumor and normal tissues. (Gao, X et al., OncoTargets and Therapy, 2019)
(D) Heatmap depicting the differential methylation of miRNA-encoding genes. (E) Volcano plot illustrating the differential expression of reads between tumor and normal tissues. (Gao, X et al., OncoTargets and Therapy, 2019)