Circular RNAs (circRNAs) modulate the expression of target genes through various mechanisms, including serving as microRNA (miRNA) sponges to influence RNA stability and translation, interacting with proteins to regulate their functions, encoding proteins, and engaging with DNA or transcription factors to control transcription. This article delves into the mechanisms by which circRNAs influence transcription.
Mechanisms of circRNA-Mediated Transcriptional Regulation
The promoter region is one of the most comprehensively investigated areas in transcriptional regulation. CircRNAs can influence the transcription of their host genes either positively or negatively. This regulation is achieved by interacting with RNA polymerase II (Pol II), recruiting proteins, or forming R-loops within the transcriptional regulatory regions of their target genes.
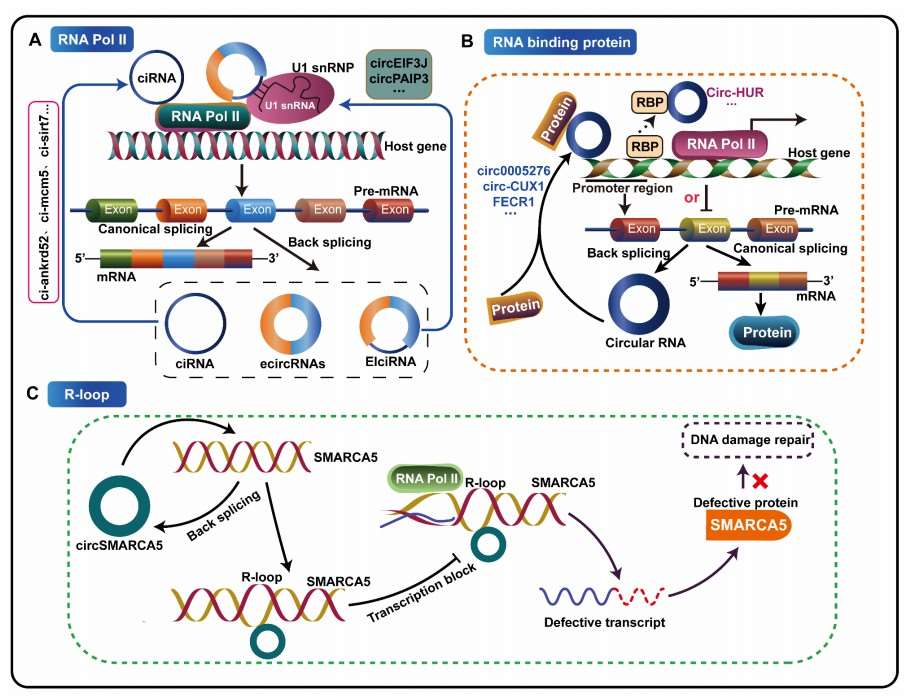 Figure 1 CircRNAs regulate host gene expression at the transcriptional level.
Figure 1 CircRNAs regulate host gene expression at the transcriptional level.
Interaction with Pol II to Enhance Host Gene Transcription
The enhancement of host gene transcriptional activity facilitated by poly II involves specific intronic ciRNAs, namely ci-ankrd52, ci-mcm5, and ci-sirt7. These ciRNAs are predominantly localized at the transcription sites of their host genes, which coincide with regions associated with RNA Polymerase II-mediated transcription elongation. Acting as positive transcriptional regulators, they significantly augment the expression levels of their host genes.
In addition, two exonic-intronic circular RNAs (EIciRNAs), circEIF3J and circPAIP2, interact with RNA Polymerase II, U1 snRNP, and the promoters of their host genes. Through the formation of positive feedback loops in a cis-acting manner, these EIciRNAs further enhance the transcriptional levels of their respective host genes.
The Recruitment of Proteins Modulating Host Gene Expression
Certain circRNAs possess highly specific protein-binding sites, enabling them to function as protein decoys, scaffolds, or recruiters. By attracting one or multiple proteins to specific regions of target promoters, these circRNAs can modulate the transcriptional activation and expression of host genes. The types of proteins recruited include RNA-binding proteins (RBPs), DNA demethylases, and DNA methyltransferases.
Activation of Host Gene Transcription via Protein Recruitment
For instance, circ-CUX1 can bind to EWS RNA-binding protein 1 (EWSR1), facilitating the interaction of EWSR1 with MYC-associated zinc finger protein (MAZ). This interaction induces the transcriptional activation of MAZ, thereby regulating the transcription of the host gene CUX1 and other related genes. This regulatory mechanism promotes aerobic glycolysis and the malignant progression of neuroblastoma.
Another example is circRNA FECR1, which recruits the DNA demethylase TET1 to the promoter of its host gene FLI1, inducing DNA demethylation. Furthermore, circRNA FECR1 binds to and downregulates the DNA methyltransferase DNMT1, resulting in hypomethylation of promoter CpG islands and activation of FLI1 transcription. This process enhances the invasive capacity of breast cancer cells.
Inhibition of Host Gene Transcription Through RBP Sequestration
circ-HUR exemplifies the inhibitory function of circRNAs. It interacts with the RGG domain of CCHC-type zinc finger nucleic acid-binding protein (CNBP), preventing CNBP from binding to the HuR promoter. This interaction inhibits the transcription of HuR, leading to the downregulation of its host gene HuR. As a result, the growth and invasiveness of gastric cancer are suppressed both in vitro and in vivo.
Regulation of Host Gene Expression via R-loop Formation
An R-loop is a specialized chromatin structure composed of an RNA-DNA hybrid and displaced single-stranded DNA. R-loops often form as a result of RNA polymerase stalling or defects in RNA biogenesis. These structures can interfere with DNA replication, repair, and transcription. CircRNAs may enhance the splicing efficiency of homologous exon-deficient mRNA by hybridizing with DNA or forming R-loops. This not only affects the abundance of linear transcripts but also creates mRNA traps, pausing transcription and improving the recruitment of splicing factors.
In summary, circRNAs play versatile roles in regulating host gene expression through protein recruitment, RBP sponging, and R-loop formation. Their ability to modulate key epigenetic and transcriptional processes underscores their significance in tumorigenesis and presents novel therapeutic opportunities for treating malignancies.
Investigating the Transcriptional Regulation of circRNA: A Case Study
Background and Context
The study under discussion, published in Molecular Cancer, a highly respected journal with a significant impact factor, delves into the molecular mechanisms underlying gemcitabine (GEM) resistance in pancreatic ductal adenocarcinoma (PDAC). This resistance represents a critical challenge in the therapeutic management of PDAC, a notoriously aggressive malignancy with limited treatment options. In particular, the focus is on circRN), which has garnered substantial attention in recent years due to its involvement in cancer progression, chemoresistance, and regulatory functions at the transcriptional and post-transcriptional levels.
In this study, hsa_circ_0007919 is identified as a key player in GEM resistance through its modulation of the DNA damage response (DDR) and repair pathways. The upregulation of hsa_circ_0007919 in GEM-resistant PDAC tissues suggests a functional role in maintaining cellular survival under chemotherapy stress. The investigation outlines how hsa_circ_0007919 recruits transcription factors such as FOXA1 and TET1, driving the expression of LIG1 (Ligase I), a gene integral to DNA repair mechanisms. The study emphasizes the importance of this regulatory axis in enabling PDAC cells to evade apoptosis and sustain proliferation despite DNA damage induced by GEM.
Study Design and Methodology
The study employed a multifaceted approach to elucidate the role of hsa_circ_0007919 in the transcriptional regulation of LIG1, using state-of-the-art technologies and experimental designs:
Next-Generation Sequencing (NGS): Initial screening through RNA sequencing revealed a marked upregulation of hsa_circ_0007919 in GEM-resistant PDAC tissues and cell lines, providing a foundation for subsequent functional analyses.
Cellular Assays: Functional assays were conducted to evaluate the impact of hsa_circ_0007919 on cell proliferation, apoptosis, and sensitivity to GEM. Silencing hsa_circ_0007919 reduced cell survival and increased chemosensitivity, while its overexpression conferred resistance.
RNA Sequencing (RNA-seq): Correlational analysis between hsa_circ_0007919 and LIG1 expression levels was performed, establishing a positive association. This suggested that hsa_circ_0007919 enhances LIG1 transcription, thus promoting GEM resistance through enhanced DNA repair activity.
DNA Damage Response Pathway Activation: Experimental data indicated that hsa_circ_0007919 induces LIG1 expression, leading to the activation of DDR pathways, including base excision repair (BER), mismatch repair (MMR), and nucleotide excision repair (NER). This facilitated the efficient repair of GEM-induced DNA lesions, thereby sustaining PDAC cell viability.
Subcellular Localization Studies (FISH/Nuclear-Cytoplasmic Fractionation): Fluorescence in situ hybridization (FISH) and nuclear-cytoplasmic fractionation assays demonstrated that hsa_circ_0007919 is predominantly localized in the nucleus, reinforcing its role in transcriptional regulation.
Protein Interaction Predictions and Database Analysis: Databases such as circAtlas, STRING, and SPP were utilized to predict potential interactions between hsa_circ_0007919, FOXA1, TET1, and the LIG1 promoter. These analyses provided a bioinformatic basis for further validation experiments.
Co-Immunoprecipitation (Co-IP) and RIP/ChIRP Assays: FOXA1 and TET1 were shown to physically interact, as demonstrated by Co-IP experiments. RIP and ChIRP assays confirmed that hsa_circ_0007919 binds to both FOXA1 and TET1, suggesting a direct role in recruiting these factors to the LIG1 promoter.
CpG Island Methylation and Transcription Factor Binding: Using JASPAR and MethPrimer 2.0, the study predicted FOXA1 binding to CpG islands in the LIG1 promoter. Chromatin immunoprecipitation (ChIP) assays verified the binding of FOXA1 and TET1 to the -1273 to -1411 region of the LIG1 promoter, correlating with decreased promoter methylation and increased transcriptional activation.
Luciferase Reporter Assays: To confirm the transcriptional activation of LIG1, luciferase reporter constructs containing the LIG1 promoter were used. The results demonstrated that hsa_circ_0007919, in conjunction with FOXA1 and TET1, significantly enhanced LIG1 promoter activity.
From the above research ideas, it can be seen that the most critical technologies for studying transcriptional regulation of circRNA or other RNA (such as lncRNA) are as follows:
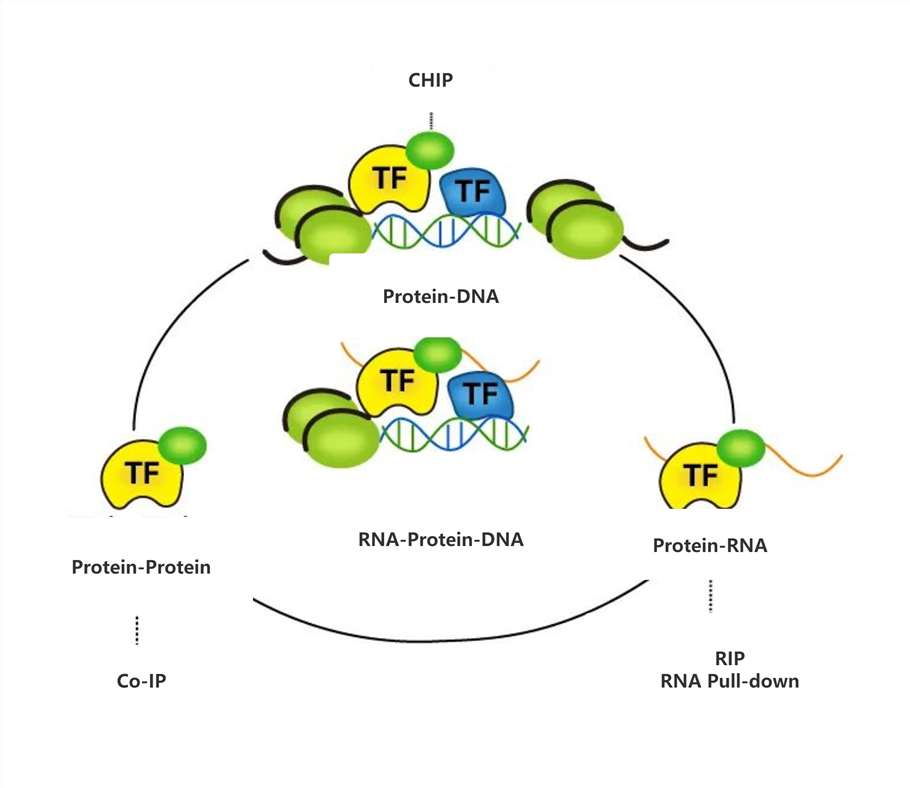 Figure 2 Technologies for studying transcriptional regulation of circRNA
Figure 2 Technologies for studying transcriptional regulation of circRNA
Conclusions and Implications
The findings of this study underscore the critical role of hsa_circ_0007919 in modulating GEM resistance in PDAC by regulating the transcriptional activation of LIG1 through its interaction with FOXA1 and TET1. By reducing the methylation of the LIG1 promoter, hsa_circ_0007919 facilitates the upregulation of DNA repair pathways, thereby enabling cancer cells to resist chemotherapy-induced cytotoxicity.
These results provide significant insights into the molecular mechanisms of chemotherapy resistance and suggest that targeting the hsa_circ_0007919-FOXA1-TET1-LIG1 axis may represent a novel therapeutic strategy for overcoming GEM resistance in PDAC. Furthermore, this study highlights key techniques and approaches—such as NGS, ChIP, and luciferase reporter assays—that are pivotal for exploring the transcriptional regulation of circRNAs and other non-coding RNAs.
This research also opens avenues for further investigation into the broader role of circRNAs in transcriptional regulation, DNA repair mechanisms, and cancer chemoresistance. The integration of advanced molecular techniques, including CRISPR-based genome editing, might further elucidate the functional relevance of circRNA-driven transcriptional networks in oncogenesis and therapy resistance.
In conclusion, this study exemplifies a comprehensive approach to understanding the molecular intricacies of chemoresistance, laying the groundwork for future therapeutic developments aimed at improving the efficacy of chemotherapeutic regimens in PDAC and other cancers characterized by resistance to conventional therapies.
References:
- Wei J, et al. Understanding the roles and regulation patterns of circRNA on its host gene in tumorigenesis and tumor progression. J Exp Clin Cancer Res. 2023;42(1):86.
- Zhang Y, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792-806.
- Li Z, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256-64.
- Li H, et al. Therapeutic targeting of circCUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol Med. 2019;11(12):e10835.
- Chen N, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19(1):218.
- Yang F, et al. Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol Cancer. 2019;18(1):158.
- Xu L, et al. hsa_circ_0007919 induces LIG1 transcription by binding to FOXA1/TET1 to enhance the DNA damage response and promote gemcitabine resistance in pancreatic ductal adenocarcinoma. Mol Cancer. 2023;22(1):195.


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1 CircRNAs regulate host gene expression at the transcriptional level.
Figure 1 CircRNAs regulate host gene expression at the transcriptional level.  Figure 2 Technologies for studying transcriptional regulation of circRNA
Figure 2 Technologies for studying transcriptional regulation of circRNA 