While the soil harbors a plethora of phages, a significant portion of their host organisms remains unidentified, hindering genomic characterization. A recent study has addressed this knowledge gap by employing advanced high-throughput chromosome conformation capture (Hi-C) technology. This innovative approach allows for the direct capture of phage-host relationships in soil, providing empirical evidence for the role of phages in shaping bacterial population dynamics within this environment.
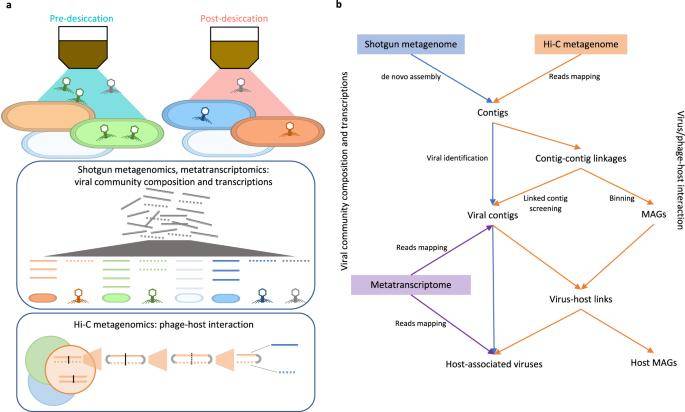 Schematic of the experimental design and data analysis workflow. (Wu et al., 2023)
Schematic of the experimental design and data analysis workflow. (Wu et al., 2023)
Identification of Infections in the Soil Microbiome Using Hi-C Metagenome Sequencing
The study yielded 479 viral operational taxonomic units (vOTUs), all identified as phages. Approximately half of these vOTUs defied classification, while the remainder were attributed to the Caudoviticetes class. A viral tree with three major branches effectively portrayed the genome-wide similarity of vOTUs in each sample. Through Hi-C metagenomic analysis, 19 phages were conclusively linked to distinct bacterial metagenome-assembled genome (MAG) hosts.
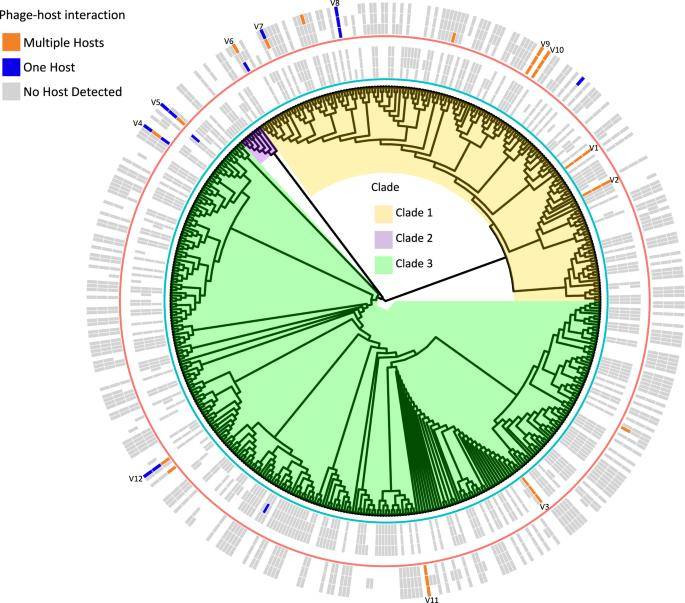 Soil phage–host interactions revealed using Hi–C metagenomics. (Wu et al., 2023)
Soil phage–host interactions revealed using Hi–C metagenomics. (Wu et al., 2023)
These host-associated phages constituted a notable portion, ranging from 5.3% to 15.0%, of the total phage sequence abundance observed in the samples. Furthermore, the study uncovered 124 CRISPR spacer sequences within CRISPR arrays in MAG, demonstrating matches to phages and revealing 121 unique phage-host junctions.
Hi-C and Metatranscriptome Reveal More Lysogeny Following Soil Drying
The study compared results before and after a two-week drying incubation, simulating the summer drying typical of arid prairie soils. The findings unveiled significant shifts in the soil phage community due to soil drying, with only 18.0% of total viral operational taxonomic units (vOTUs) detected in both pre- and post-drying soils.
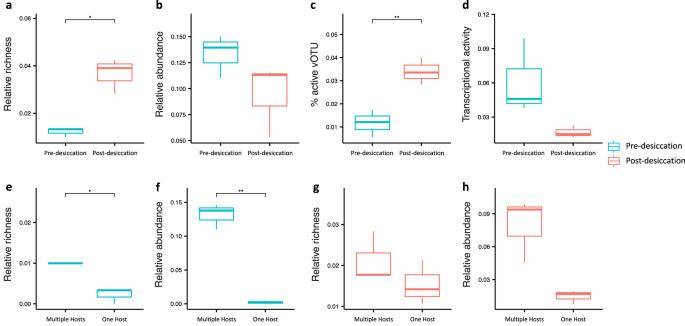 Richness, abundance, and transcriptional activities of host-associated vOTUs. (Wu et al., 2023)
Richness, abundance, and transcriptional activities of host-associated vOTUs. (Wu et al., 2023)
The impact of soil drying on host-associated phage communities was noteworthy, as evidenced by Hi-C metagenome analysis. Post-drying soil exhibited a significantly higher proportion of host-associated vOTUs compared to pre-drying soil (p < 0.05). However, the relative abundance of these host-associated vOTUs did not show a significant difference between pre- and post-drying soils (p = 0.18).
Metatranscriptomic data provided additional insights, revealing a higher percentage of transcriptionally active host-associated vOTUs after soil drying. However, despite this increase, the average transcriptional activity of these vOTUs showed a decline after soil drying, as indicated by a lower percentage of transcripts aligned to host-associated vOTUs.
Hi-C Reconstruction of Soil Phage Host Infection Network
Metagenome-assembled genomes (MAGs) identified as phage hosts through Hi-C sequencing encompassed various bacterial phyla, including Acidobacteria, Actinobacteria, Chlamydiae, Bacteriophages, Gemmatimonadota, and Proteobacteria. Each MAG associated with one or more phages formed a distinct host group.
In pre-drying soil, two of the five host-associated viral operational taxonomic units (vOTUs) were linked to a single host MAG, while the remaining three vOTUs were associated with multiple hosts (V1, V2, and V3). VOTUs connected to multiple hosts exhibited higher richness and abundance compared to those linked to a single host. Certain hosts were prominently concentrated in the bacterial community co-occurrence network, indicating that phage infection significantly influences interactions within the soil-bacterial community.
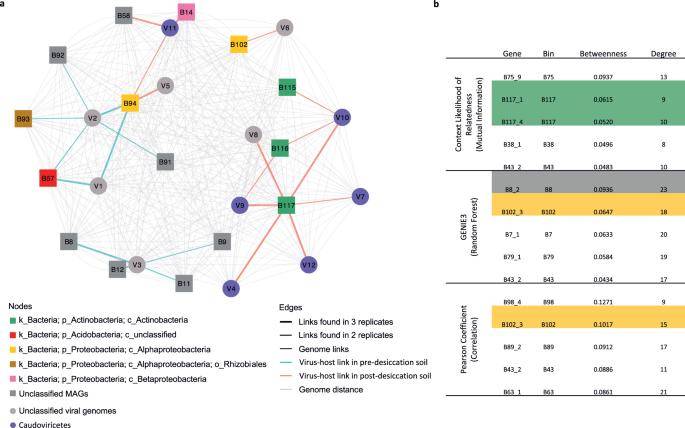 Phage–host infection network and community co-occurrence analysis. (Wu et al., 2023)
Phage–host infection network and community co-occurrence analysis. (Wu et al., 2023)
The community co-occurrence network was utilized to identify microbial members with high centrality, signifying their extensive connections to other species or their pivotal positions in the network. Results indicated that MAGs serving as phage hosts were among the most central nodes in the network. Interestingly, abundance did not emerge as the primary factor driving centrality in the host co-abundance network.
Phage Infection Regulates Host Population Dynamics
The study noted a rise in the average viral copy number (VPH) per host and a reduction in viral transcriptional activity following a two-week soil-drying incubation, indicating an uptick in lysogenic infection. Additionally, soil drying induced changes in the observed phage host range. Notably, the substantial negative correlation between VPH and host abundance before drying implies that heightened lysogenic infections result in increased host mortality, exerting a negative impact on host abundance.
Reference:
- Wu, R., Davison, M.R., Nelson, W.C. et al. Hi-C metagenome sequencing reveals soil phage–host interactions. Nat Commun 14, 7666 (2023).
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Schematic of the experimental design and data analysis workflow. (Wu et al., 2023)
Schematic of the experimental design and data analysis workflow. (Wu et al., 2023) Soil phage–host interactions revealed using Hi–C metagenomics. (Wu et al., 2023)
Soil phage–host interactions revealed using Hi–C metagenomics. (Wu et al., 2023) Richness, abundance, and transcriptional activities of host-associated vOTUs. (Wu et al., 2023)
Richness, abundance, and transcriptional activities of host-associated vOTUs. (Wu et al., 2023) Phage–host infection network and community co-occurrence analysis. (Wu et al., 2023)
Phage–host infection network and community co-occurrence analysis. (Wu et al., 2023)