Understanding the Dynamic World of Gene Translation
Translome, a term encompassing all components directly involved in the translation process, opens a window into the intricate world of molecular biology. It includes, but is not limited to, the core players like ribosomes, in-translated mRNAs (RNC-mRNAs), tRNAs, and a variety of translation factors. Additionally, it extends its reach to regulatory RNAs, such as miRNAs and lncRNAs, and even de novo peptide chains. Translatomics, a burgeoning field, investigates the composition and dynamic patterns of these essential components.
Please refer to our article What is Translatomics?
Translational Insights Under Abiotic Stress
Plant life faces a barrage of challenges, with abiotic stresses like extreme temperatures, light intensity variations, drought, salinity, and waterlogging. These environmental factors significantly impact gene expression in plants. Understanding how root systems navigate these challenges under varying irrigation and rainfall conditions is pivotal in developing climate-resilient crops.
Recent studies have shed light on the translatome and chromatin profiles of rice root cells under different environmental conditions. Submergence, for instance, dampens the DNA synthesis of cell cycle-related genes during proliferation. It also impacts genes associated with growth hormone signaling, the biological clock, and small RNA regulation in ground tissues. Meanwhile, corky matter biosynthesis, iron transport proteins, and nitrogen assimilation in endosperm/extrinsic cells, all water availability-dependent, experience regulatory changes under submerged conditions. These findings highlight the intricate web of gene regulation responding to abiotic stress and identify critical transcription factors driving responses to water deficits and xylem development.
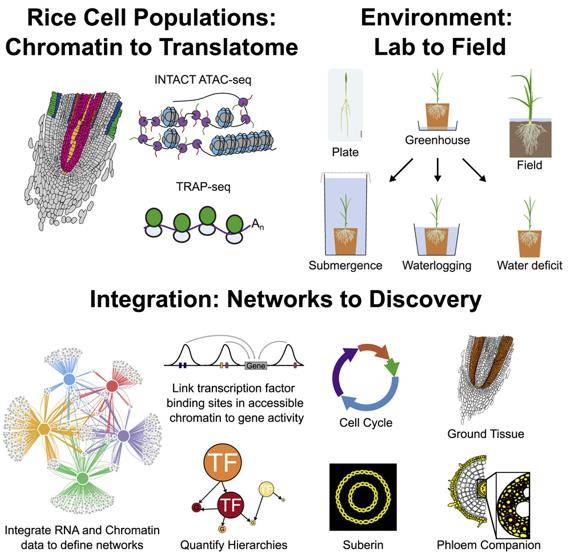 An atlas of gene activity of root cell populations of rice in varied agronomically relevant environments. (Reynoso et al., 2022)
An atlas of gene activity of root cell populations of rice in varied agronomically relevant environments. (Reynoso et al., 2022)
Unveiling Translatome Responses to Biotic Stress
In the face of biotic threats, plants engage in transcriptional reprogramming to protect themselves. However, much remains undiscovered about translational reprogramming during plant immune responses. Researchers are now applying cutting-edge techniques like Ribo-Seq and RNA sequencing to decode the translational responses of host plants to infections.
For example, a study examined the porcine reproductive and respiratory syndrome virus (PRRSV) translatome. This research revealed gene expression strategies that enable the virus to exert translational control during its replication cycle. The study found shifting efficiencies at specific loci and identified additional viral ORFs that may be facilitated by new viral transcripts. These discoveries provide deeper insights into the intricate mechanisms of viral gene expression.
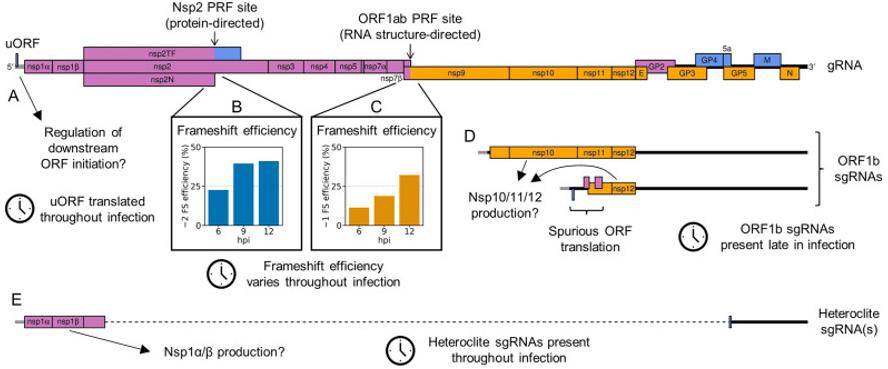 Schematic summary of non-canonical mechanisms of porcine reproductive and respiratory syndrome virus (PRRSV) gene expression regulation. (Cook et al., 2022)
Schematic summary of non-canonical mechanisms of porcine reproductive and respiratory syndrome virus (PRRSV) gene expression regulation. (Cook et al., 2022)
Translational Control in Development
Translational control is not limited to stress responses. It plays a crucial role in regulating gene expression during development, facilitating tissue-specific gene product synthesis in both plants and animals.
Recent investigations have explored the full spectrum of mRNAs translated post-fertilization by analyzing polysomes and sequencing. Surprisingly, only a fraction of maternal mRNAs are selectively recruited to polysomes. This recruitment, independent of mRNA abundance, is predominantly composed of mRNAs linked to specific functional classes associated with cell cycle transitions and later developmental events. Intriguingly, fertilization might involve an initiation model that doesn't rely on the mTOR pathway, as suggested by the variable response of newly translated mRNAs to mTOR pathway impairment.
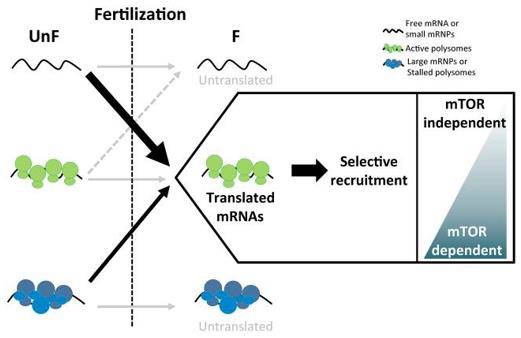 Model of polysomal recruitment dynamics upon fertilization in sea urchin. (Chassé et al., 2018)
Model of polysomal recruitment dynamics upon fertilization in sea urchin. (Chassé et al., 2018)
Translocomic Insights into Organelle Genes
Organelles like chloroplasts and mitochondria are pivotal in fundamental plant processes such as photosynthesis and respiration. These tiny powerhouses contain their own miniature genomes, comprising only a handful of genes. Chloroplasts, for instance, respond to temperature changes, impacting plant cold tolerance.
Research has delved into cold domestication in tobacco seedlings exposed to low temperatures, scrutinizing chloroplast genome copy numbers, transcript accumulation, translation, photosynthesis, cellular physiology, and metabolic responses. The findings reveal significant cold-induced translational regulation at the initiation and elongation levels, with minimal changes in transcript levels. This highlights the previously underestimated role of chloroplast genes in plant cold tolerance.
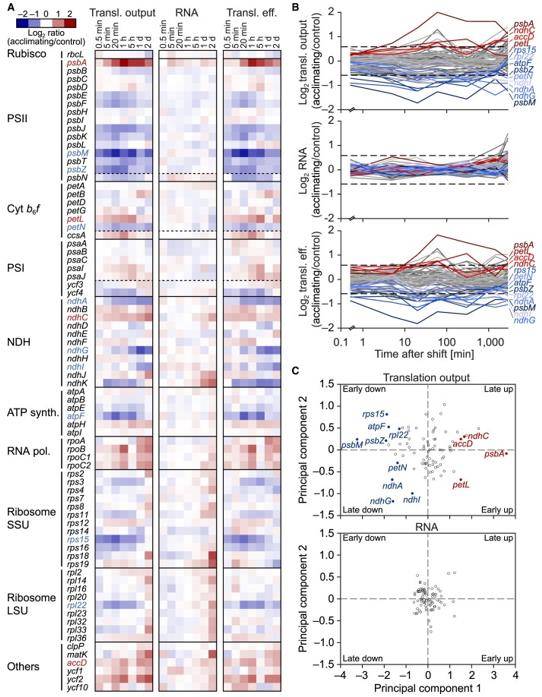 Extensive translational regulation in tobacco chloroplasts during acclimation to low temperature. (Gao et al., 2022)
Extensive translational regulation in tobacco chloroplasts during acclimation to low temperature. (Gao et al., 2022)
Translome Technology is rapidly expanding our understanding of the complex world of gene translation in both healthy and stressed plant systems. These advances in research are opening up new avenues for the development of resilient crops and deepening our understanding of plant biology at the molecular level.
References:
- Reynoso, Mauricio A., et al. "Gene regulatory networks shape developmental plasticity of root cell types under water extremes in rice." Developmental Cell 57.9 (2022): 1177-1192.
- Cook, Georgia M., et al. "Ribosome profiling of porcine reproductive and respiratory syndrome virus reveals novel features of viral gene expression." Elife 11 (2022): e75668.
- Chassé, Héloïse, et al. "Translatome analysis at the egg-to-embryo transition in sea urchin." Nucleic Acids Research 46.9 (2018): 4607-4621.
- Gao, Yang, et al. "Chloroplast translational regulation uncovers nonessential photosynthesis genes as key players in plant cold acclimation." The Plant Cell 34.5 (2022): 2056-2079.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 An atlas of gene activity of root cell populations of rice in varied agronomically relevant environments. (Reynoso et al., 2022)
An atlas of gene activity of root cell populations of rice in varied agronomically relevant environments. (Reynoso et al., 2022) Schematic summary of non-canonical mechanisms of porcine reproductive and respiratory syndrome virus (PRRSV) gene expression regulation. (Cook et al., 2022)
Schematic summary of non-canonical mechanisms of porcine reproductive and respiratory syndrome virus (PRRSV) gene expression regulation. (Cook et al., 2022)  Model of polysomal recruitment dynamics upon fertilization in sea urchin. (Chassé et al., 2018)
Model of polysomal recruitment dynamics upon fertilization in sea urchin. (Chassé et al., 2018) Extensive translational regulation in tobacco chloroplasts during acclimation to low temperature. (Gao et al., 2022)
Extensive translational regulation in tobacco chloroplasts during acclimation to low temperature. (Gao et al., 2022)