With the advent of single-cell RNA sequencing (scRNA-seq) technology, significant strides have been made in the field of drug development. This approach, by meticulously profiling the gene expression landscapes of individual cells, offers a novel, efficient, and precise method for investigating drug mechanisms of action. Through scRNA-seq, we gain deep insights into the specific impacts of drugs on processed cells and tissues, thereby enhancing our understanding of the molecular mechanisms underlying drug action. The widespread application of this technology holds great promise for vastly improving the efficiency of drug screening and development, ultimately providing more precise and effective therapeutic solutions for disease treatment.
Recently, Professor Edgardo Ferran and his team from the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI) in the United Kingdom published a comprehensive review titled "Applications of single-cell RNA sequencing in drug discovery and development" in the prestigious academic journal Nature Reviews Drug Discovery. This review extensively surveys the broad applications of scRNA-seq technology throughout the drug discovery and development pipeline, spanning from target identification to clinical decision-making. Furthermore, the article delves into the challenges associated with implementing scRNA-seq technology in the pharmaceutical industry, offering valuable insights and guidance for advancing research in related fields.
Drug development has historically been a time-consuming, costly, and high-risk process. Despite promising preclinical potential exhibited by many drugs, their success rates often falter upon entering clinical trials due to various complicating factors. Among these, limitations in our understanding of human biology—especially in-depth insights into disease mechanisms, effective identification of therapeutic targets, and grasp of individual differences in disease response—pose significant hurdles.
Before the advent of single-cell (SC) technologies, researchers relied heavily on analyses of large sample sizes of population cells and tissues. However, the advent of next-generation sequencing (NGS), particularly breakthroughs in single-cell sequencing, has greatly expanded the depth and breadth of genomic and biomedical research. Leveraging these cutting-edge technologies, we can achieve deep whole-genome (DNA-seq), whole-epigenome, and whole-transcriptome sequencing at the single-cell level, thereby gaining more precise insights into the mysteries of life.
Since 2009, scRNA-seq technology has made significant strides and found widespread application in the pharmaceutical industry. The proliferation and maturation of this technology provide robust solutions to key challenges in drug discovery and development, thereby advancing innovation in the medical field.
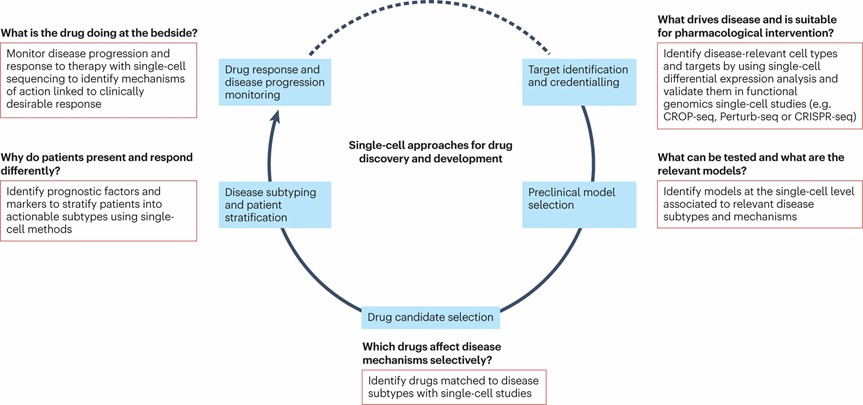 Figure 1. How scRNA-seq can inform decision making during drug discovery and development
Figure 1. How scRNA-seq can inform decision making during drug discovery and development
A typical scRNA-seq workflow encompasses three core stages: library construction, preprocessing, and post-processing. During the library construction stage, procedures primarily involve the isolation of individual cells or nuclei, mRNA capture, and subsequent sequencing operations (Figure 2).
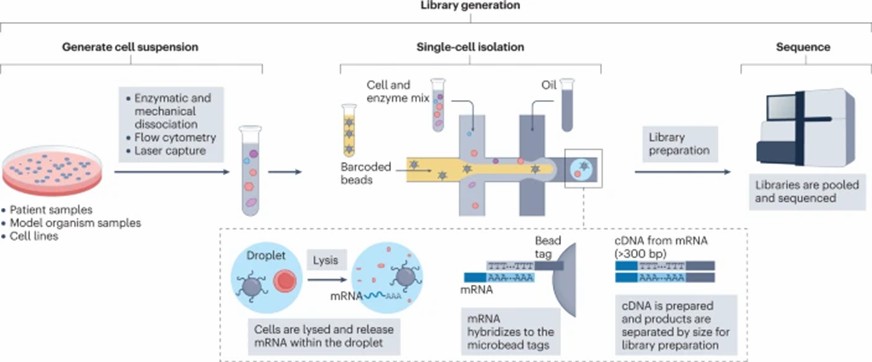 Figure 2. ScRNA-seq library generation process
Figure 2. ScRNA-seq library generation process
scRNA-Seq in Drug Development
scRNA-seq has been extensively employed throughout the drug development pipeline. By accurately classifying cellular compositions and states at subtypes, this technology enhances our understanding of disease fundamentals, thereby providing robust guidance for the discovery of new cellular and molecular targets. Additionally, scRNA-seq demonstrates immense potential in target identification and validation, facilitating the identification of preclinical models closely associated with specific disease subtypes.
During the critical phase of candidate drug selection, scRNA-seq sequencing technology offers profound insights into cell-type-specific compound interactions, off-target effects, and heterogeneous responses, thereby providing a scientific basis for drug development decisions. Throughout clinical development, this technology plays an indispensable role by aiding researchers in identifying biomarkers for patient stratification, elucidating drug mechanisms of action and resistance, monitoring drug responses, and disease progression, thereby ensuring the safety and efficacy of drugs.
Notably, scRNA-seq technology also presents new opportunities for characterizing and improving emerging fields such as engineered biological products and cell therapies, continuously driving innovation and development in these areas. With ongoing technological advancements and expanding application domains, scRNA-seq is poised to play an increasingly pivotal role in drug development.
Understanding Disease
Many complex diseases involve interactions among multiple cell types, and scRNA-seq technology has significantly advanced our understanding of their underlying nature with its high-resolution capabilities. Utilizing scRNA-seq, we can precisely capture differences between cell types and changes in cell phenotypes, which often represent significant features of pathological states. Moreover, the objective perspective of scRNA-seq allows us to detect rare cell types that may play crucial roles in disease biology processes.
Currently, this technology is providing profound insights into potential disease mechanisms, thereby facilitating exploration of novel therapeutic strategies. Whether in cancer, neurodegenerative diseases, inflammation, autoimmune disorders, or infectious diseases, scRNA-seq demonstrates unique advantages and potentials (Figure 3). We believe that with ongoing development and refinement, this technology will continue to bring about breakthroughs and advancements in disease research and treatment.
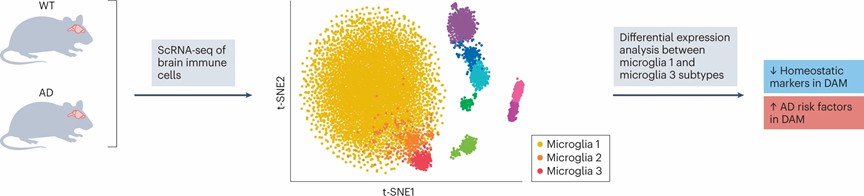 Figure 3. scRNA-seq in disease understanding
Figure 3. scRNA-seq in disease understanding
Target Discovery in Drug Development
In oncology, single-cell methods have gained prominence for their pioneering role in identifying precise therapeutic strategies directly targeting cancer cells. Concurrently, scRNA-seq technology, leveraging its unique advantages, extends into other therapeutic domains, bolstering target discovery efforts.
During target validation, ensuring the credibility of gene targets necessitates comprehensive consideration of evidence from disease biology, target biology, druggability, and genetic studies. Furthermore, assessing the translational efficacy of research models allows for a deeper analysis of potential disparities between models and disease biology or therapeutic targets. In this process, scRNA-seq data plays a pivotal role by providing valuable insights for target validation.
In preclinical studies, scRNA-seq technology expands the evaluation focus beyond traditional batch or averaged approaches to precise measurements of cellular composition, tissue heterogeneity, and rare cell phenotypes. This shift significantly enhances the accuracy in assessing model fidelity or patient relevance, aiding in the selection of models that best meet clinical needs and thereby improving the success rate of clinical translation.
Drug Screening and Mechanism of Action (MoA) Analysis
In the field of drug discovery, traditional high-throughput screening (HTS) methods often rely on relatively coarse metrics such as cell viability or proliferation, or highly specific evaluations based on the expression of particular markers. However, with technological advancements, the latest HTS methods have begun to integrate scRNA-seq technology, enabling more precise and comprehensive assessments.
Traditional HTS typically involves extensive compound libraries but is limited by testing conditions and methodologies, usually employing a single dose under highly restricted biological environments. In contrast, novel HTS approaches leverage the rich information provided by single-cell gene expression profiles, allowing for the simultaneous testing of multiple doses and extensive evaluation under diverse biological conditions. This improvement significantly enhances the potential and advantages of new HTS methods in studying drug mechanisms of action (Figure 4).
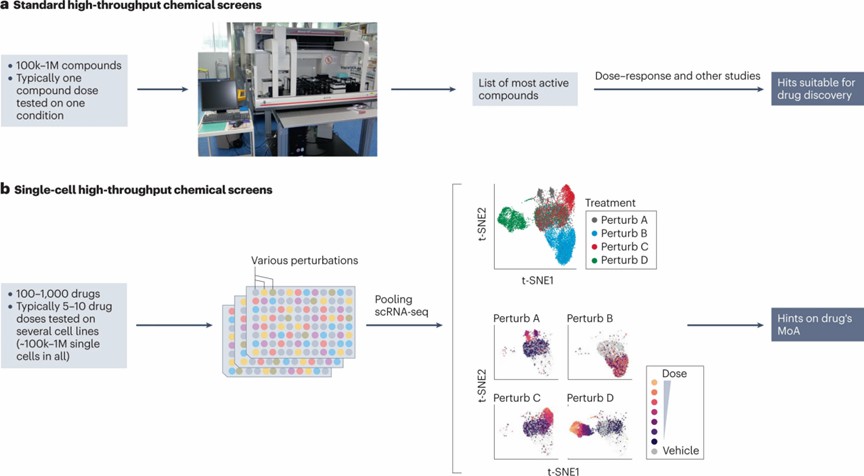 Figure 4. Single-cell high-throughput screening
Figure 4. Single-cell high-throughput screening
Biomarkers and Patient Stratification
Recently, scRNA-seq has played a critical role in precisely defining prognostic biomarkers for colorectal cancer (CRC) (Figure 5). Additionally, scRNA-seq has been widely used to characterize mechanisms of chemotherapy resistance in cancers, as demonstrated in studies on high-grade serous ovarian cancer (HGSOC). Currently, numerous studies are focused on applying scRNA-seq to analyze diseased tissues to uncover biomarkers that predict drug response or resistance. Despite these advancements, challenges and limitations remain in translating these findings into clinical applications.
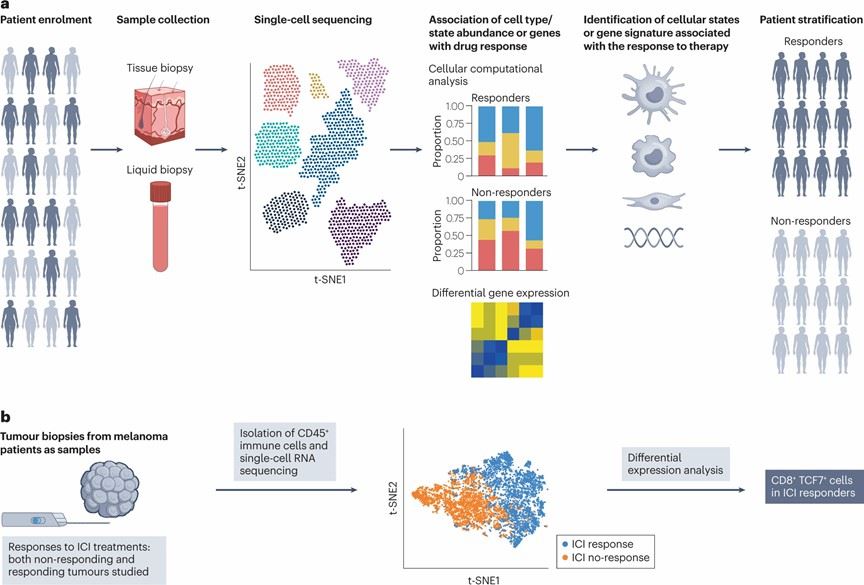 Figure 5. Biomarker Discovery and Patient Stratification.
Figure 5. Biomarker Discovery and Patient Stratification.
Monitoring Drug Response and Disease Progression
In recent years, single-cell sequencing methods have had a significant impact on clinical decision-making for monitoring therapeutic responses, particularly in oncology. Minimal residual disease (MRD) serves as a crucial indicator for evaluating the presence of residual cancer cells during or after treatment, and remains a core standard for measuring drug response efficacy. Compared to traditional MRD assessment methods, the current approach, which integrates single-cell mutation analysis, enables precise subclonal-level evaluations at lower detection limits and comprehensively analyzes the dynamic evolution of subclones throughout treatment.
The application of single-cell mutation analysis has not only significantly enhanced the sensitivity and specificity of MRD detection but also effectively identifies resistant subclones that may lead to disease relapse. Notably, some relapse risks associated with MRD arise from persistent cells that respond to treatment and are induced by non-genetic adaptive mechanisms. Our current understanding of these mechanisms is still limited.
To investigate these rare, transiently drug-resistant persistent cells, researchers have developed a high-complexity lentiviral barcode library known as Watermelon. This library can simultaneously track the clonal lineage, proliferation status, and transcriptomic profile of individual cells during drug treatment, providing a powerful tool for elucidating the mechanisms of cellular drug resistance.
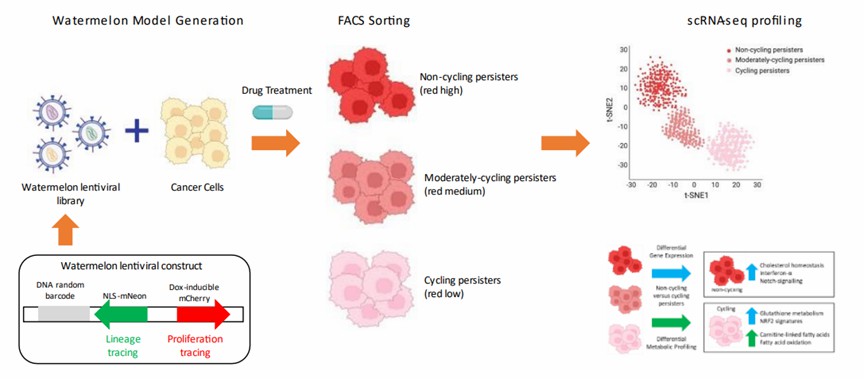 Figure 6. Monitoring drug response using the Watermelon library
Figure 6. Monitoring drug response using the Watermelon library
Current Challenges
Despite the significant advancements in drug discovery and development facilitated by single-cell sequencing technologies, particularly scRNA-seq, along with associated computational tools and public data resources, the field still faces numerous challenges. To fully harness the transformative potential of these technologies, the industry must confront and address these challenges by adapting its infrastructure and workflows. Additionally, given that the volume of publicly available scRNA-seq data far exceeds the output of any single pharmaceutical company, effectively integrating this vast amount of data remains a daunting task.
Moreover, due to the sample requirements and cost factors involved in generating scRNA-seq data, this technology is not yet poised to replace large-scale molecular profiling of early discovery or clinical samples. Consequently, a key issue that needs to be addressed is how to effectively integrate scRNA-seq data with large-scale molecular profiling datasets.
To tackle these challenges, several critical factors must be considered:
- Careful planning and rigorous execution of study design and implementation strategies.
- Ensuring broad accessibility of scRNA-seq data to promote data sharing and collaboration.
- Enhancing data interoperability and reusability to foster cross-disciplinary and cross-platform collaborative research.
In summary, single-cell sequencing technologies, particularly scRNA-seq, along with their associated computational tools and public data resources, are indispensable for deepening our understanding of disease mechanisms and precisely identifying the most therapeutically promising cellular or molecular targets. Additionally, scRNA-seq technology aids in selecting appropriate preclinical disease models and provides novel perspectives for elucidating drug mechanisms of action. By accelerating the discovery of new biomarkers, this technology supports precise patient stratification, enabling the development of personalized treatment plans based on prognostic or predictive responses, thus driving the advancement of personalized medicine.
References:
-
Van de Sande B, Lee J S, Mutasa-Gottgens E, et al. Applications of single-cell RNA sequencing in drug discovery and development. Nature Reviews Drug Discovery, 2023: 1-25.
- Wu H, Wang C, Wu S. Single-Cell Sequencing for Drug Discovery and Drug Development. Curr Top Med Chem. 2017;17(15):1769-1777.
- Chen H, Ye F, Guo G. Revolutionizing immunology with single-cell RNA sequencing. Cell Mol Immunol. 2019 Mar;16(3):242-249.


 Sample Submission Guidelines
Sample Submission Guidelines
 Figure 1. How scRNA-seq can inform decision making during drug discovery and development
Figure 1. How scRNA-seq can inform decision making during drug discovery and development Figure 2. ScRNA-seq library generation process
Figure 2. ScRNA-seq library generation process Figure 3. scRNA-seq in disease understanding
Figure 3. scRNA-seq in disease understanding Figure 4. Single-cell high-throughput screening
Figure 4. Single-cell high-throughput screening Figure 5. Biomarker Discovery and Patient Stratification.
Figure 5. Biomarker Discovery and Patient Stratification. Figure 6. Monitoring drug response using the Watermelon library
Figure 6. Monitoring drug response using the Watermelon library