A Comprehensive Review of RNA Sequencing Databases: Resources for Transcriptomics Research
The advent of RNA sequencing (RNA-seq) has revolutionized gene expression analysis, facilitating high-throughput insights into transcriptional landscapes across diverse biological contexts. Given the proliferation of RNA-seq data, the establishment and utilization of specialized databases are indispensable for advancing transcriptomics research. This review provides a detailed examination of RNA-seq databases, encompassing general repositories, species-specific archives, non-coding RNA collections, single-cell and spatial transcriptomics resources, and specialized databases. Emphasis is placed on the functionality, accessibility, and utility of these databases in supporting comprehensive gene expression studies.
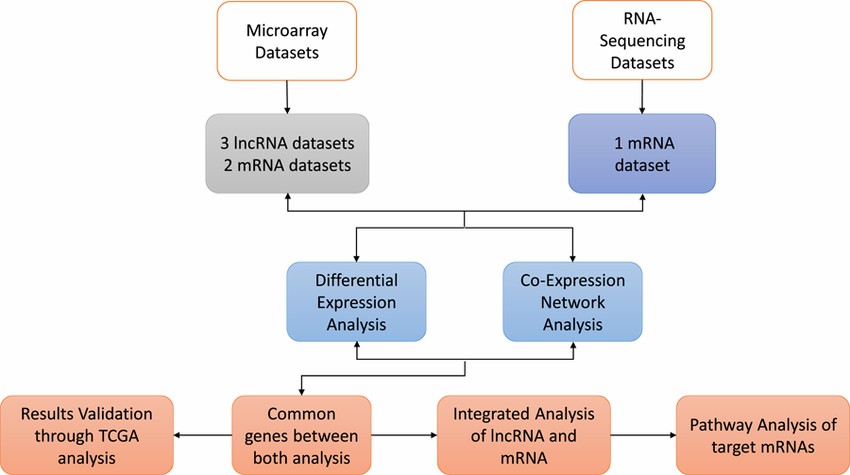 Overall Workflow of Methodology. Microarray and RNA‐Seq datasets were retrieved from the Gene Expression Omnibus (GEO) database. (Maryam Khalid et al,. 2021)
Overall Workflow of Methodology. Microarray and RNA‐Seq datasets were retrieved from the Gene Expression Omnibus (GEO) database. (Maryam Khalid et al,. 2021)
Introduction
RNA-seq technology has become a cornerstone in the investigation of gene expression, enabling comprehensive analysis of the transcriptome with unprecedented precision. The utility of RNA-seq extends across various scientific disciplines, necessitating the development of robust databases for data storage, retrieval, and analysis. This review categorizes and describes these databases, elucidating their application and significance in transcriptomics research.
General RNA-seq Databases
General RNA-seq databases provide a broad repository for RNA-seq data, accommodating diverse species and experimental conditions. They facilitate large-scale gene expression studies and cross-species comparisons.
Gene Expression Omnibus (GEO)
Description: The Gene Expression Omnibus, managed by the National Center for Biotechnology Information (NCBI), serves as a public repository for high-throughput gene expression data, including RNA-seq, microarray, and other genomic technologies.
Functions: GEO enables data submission, archiving, and retrieval, supporting extensive metadata annotation and offering robust search capabilities.
Target Audience: Researchers in genomics and molecular biology who require access to a comprehensive collection of gene expression datasets for hypothesis testing and validation.
ArrayExpress
Description: ArrayExpress, maintained by the European Bioinformatics Institute (EBI), is a curated database storing functional genomics data from high-throughput experimental techniques.
Functions: The database provides data from microarray and RNA-seq experiments, offering advanced search and analysis tools to explore gene expression patterns.
Target Audience: ArrayExpress primarily serves European researchers, though it is accessible globally for data deposition and retrieval in functional genomics.
Expression Atlas
Description: Also managed by EBI, the Expression Atlas explores gene expression across different species, tissue types, and experimental conditions.
Functions: It provides an intuitive interface for users to query gene expression data, focusing on differential expression and baseline expression levels.
Target Audience: Researchers engaged in cross-species gene expression analysis or those investigating condition-specific gene regulation.
Species-Specific and Condition-Specific RNA-seq Databases
These databases specialize in RNA-seq data for particular organisms or specific biological conditions, offering detailed expression profiles that facilitate focused research.
GTEx (Genotype-Tissue Expression)
Description: The GTEx project examines the correlation between genetic variation and gene expression across numerous human tissues.
Functions: GTEx provides extensive RNA-seq data for numerous tissues, supporting studies on gene regulation and eQTL mapping.
Target Audience: Researchers in human genetics and biomedical sciences focusing on the genetic basis of gene expression variation.
FlyBase
Description: FlyBase is dedicated to the genetics and molecular biology of Drosophila melanogaster, offering a rich repository of RNA-seq data.
Functions: It includes comprehensive gene annotations, expression data, and functional information crucial for fly genetics research.
Target Audience: Geneticists and developmental biologists utilizing Drosophila as a model organism.
WormBase
Description: WormBase provides an integrated platform for the study of the nematode Caenorhabditis elegans, encompassing extensive RNA-seq datasets.
Functions: The database supports genomic and transcriptomic data analysis, offering tools for data integration and functional annotation.
Target Audience: Researchers investigating C. elegans biology, including developmental and neurobiological studies.
ZFIN
Description: The Zebrafish Model Organism Database (ZFIN) is an essential resource for zebrafish genetics and genomics, incorporating RNA-seq data.
Functions: ZFIN offers gene expression data, genetic information, and functional annotations critical for zebrafish research.
Target Audience: Developmental biologists and geneticists focusing on zebrafish as a model system.
MaizeGDB
Description: MaizeGDB serves the maize research community, providing comprehensive genetic and RNA-seq data resources.
Functions: It includes gene expression data, genetic markers, and phenotypic information pivotal for maize genetics and breeding research.
Target Audience: Agronomists and geneticists focusing on maize improvement and functional genomics.
SoyBase
Description: SoyBase is dedicated to soybean genetics, integrating extensive RNA-seq data with genomic and phenotypic information.
Functions: The database supports advanced genomic analyses and breeding research through detailed gene expression datasets.
Target Audience: Researchers in plant genetics and agricultural science working on soybean enhancement.
RiceXPro
Description: RiceXPro provides gene expression profiles for Oryza sativa across various developmental stages and environmental conditions.
Functions: The database offers high-resolution RNA-seq data and tools for exploring gene expression in rice.
Target Audience: Plant biologists and geneticists studying rice development and stress responses.
ALDB (Arabidopsis Leaf Senescence Database)
Description: ALDB focuses on the senescence of Arabidopsis thaliana leaves, coordinating RNA-seq data for different developmental stages.
Functions: It provides gene expression information relevant to leaf aging and molecular processes in senescence.
Target Audience: Plant physiologists and molecular biologists studying senescence mechanisms.
EchinoDB
Description: EchinoDB concentrates on the sea urchin transcriptome, offering genomic and RNA-seq data sets.
Functions: The database supports analysis of gene expression during sea urchin development.
Target Audience: Evolutionary and developmental biologists using sea urchins as model organisms.
GEO Profiles
Description: As an extension of GEO, GEO Profiles facilitates the retrieval of specific gene expression profiles from stored datasets.
Functions: It allows users to search for expression data by gene, offering detailed visualization and analysis tools.
Target Audience: Researchers requiring targeted gene expression information from high-throughput experiments.
Non-coding RNA Databases
Focusing on non-coding RNAs (ncRNAs), these databases provide critical insights into the regulatory roles of these molecules in transcriptomics.
RNAcentral
Description: RNAcentral is a unified database for non-coding RNA sequences, aggregating data from multiple specialist databases.
Functions: It provides access to a broad array of ncRNA data, including sequence information and functional annotations.
Target Audience: Molecular biologists and bioinformaticians studying the roles of non-coding RNAs in gene regulation.
miRBase
Description: miRBase is the principal repository for microRNA (miRNA) sequences and annotations.
Functions: It catalogues miRNA sequences from diverse species, detailing their genomic locations and expression profiles.
Target Audience: Researchers investigating the regulatory functions of miRNAs in various biological processes.
lncRNAdb
Description: lncRNAdb provides annotations for long non-coding RNAs (lncRNAs), emphasizing their functional roles.
Functions: The database includes detailed information on lncRNA sequences, structural features, and biological functions.
Target Audience: Scientists exploring the regulatory functions and mechanisms of lncRNAs.
miRTarBase
Description: miRTarBase offers experimentally validated interactions between miRNAs and their target genes.
Functions: It provides comprehensive data on miRNA-gene interactions, supporting studies on miRNA-mediated regulation.
Target Audience: Researchers focused on understanding miRNA-target interaction networks.
Single Cell, Spatial Transcriptomics, and Epigenomics Databases
These databases support the exploration of gene expression at single-cell resolution and within spatial contexts, providing high-resolution insights into transcriptional heterogeneity.
Single Cell Portal
Description: Hosted by the Broad Institute, the Single Cell Portal contains extensive single-cell RNA-seq datasets.
Functions: It enables the visualization and analysis of single-cell gene expression data, highlighting cellular diversity and dynamics.
Target Audience: Researchers analyzing cell type-specific expression and cellular heterogeneity.
SCPortalen
Description: SCPortalen is dedicated to single-cell transcriptomics, offering a platform for data visualization and analysis.
Functions: The database facilitates the exploration of single-cell RNA-seq data, emphasizing differential gene expression.
Target Audience: Scientists investigating transcriptional diversity at the single-cell level.
EpiGenome
Description: EpiGenome integrates transcriptomic and epigenomic data, providing insights into how epigenetic changes influence gene expression.
Functions: It offers tools for analyzing the interplay between epigenetic modifications and transcriptional activity.
Target Audience: Researchers in epigenetics and gene regulation.
ASpedia
Description: ASpedia compiles data on alternative splicing events, detailing their regulatory mechanisms and functional impacts.
Functions: The database supports the investigation of splicing patterns and their influence on transcript diversity.
Target Audience: Scientists focused on RNA processing and alternative splicing regulation.
Specialized Databases
Specialized databases cater to specific areas of research, providing targeted RNA-seq data and resources to support niche fields within transcriptomics.
ImmGen (Immunological Genome Project)
Description: ImmGen offers curated RNA-seq data from murine immune cells, detailing gene expression across various immune cell types.
Functions: The database provides tools for gene expression analysis within the context of immune cell differentiation and function.
Target Audience: Immunologists studying gene regulation in immune responses.
FlyAtlas 2
Description: FlyAtlas 2 provides gene expression maps for Drosophila melanogaster, covering various tissues and developmental stages.
Functions: It supports the analysis of tissue-specific and stage-specific gene expression patterns.
Target Audience: Geneticists and developmental biologists using Drosophila as a model.
GEO
Description: As previously mentioned, GEO is a comprehensive repository for gene expression data.
Functions: It supports data submission, archival, and retrieval, facilitating broad access to high-throughput genomic data.
Target Audience: Researchers from diverse fields requiring access to extensive gene expression datasets.
Services you may interested in
The Future of RNA Sequencing Databases
The evolution of RNA-seq databases is expected to advance toward greater comprehensiveness and specialization. Emerging technologies, such as single-cell RNA sequencing, spatial transcriptomics, and in-depth studies of long non-coding RNAs, will drive the emergence of more refined databases. Additionally, as the volume of data continues to increase, effective management, integration, and analysis of these data will become pivotal research challenges.
Continued Development and Application Prospects of Databases
Data Standardization and Integration
As an increasing amount of experimental data is generated, achieving data standardization and integration across multiple databases has become a critical issue. This will facilitate cross-database comparative analysis and enhance the reusability of data.
Application of Artificial Intelligence and Machine Learning
With the incorporation of artificial intelligence (AI) and machine learning (ML) technologies, future RNA-seq databases will extend beyond mere data storage and sharing. They will offer advanced data analysis and predictive capabilities. Researchers will be able to utilize these tools to uncover novel gene expression patterns or potential biomarkers.
User-Friendliness and Visualization Tools
To enable more researchers to access and utilize these data effectively, the user interfaces of databases will become more user-friendly and provide more intuitive visualization tools. This will streamline the process of interpreting complex data and enhance research efficiency.
Diversity and Interdisciplinary Collaboration
Future databases will place greater emphasis on interdisciplinary data integration, encompassing data from fundamental biology to clinical medicine. This will foster collaboration among scientists from diverse fields and advance translational medicine.
Data Security and Privacy Protection
As the sensitivity of human genomics data increases, balancing open data sharing with personal privacy protection will remain a crucial issue. Future RNA-seq databases will further strengthen data security measures to ensure lawful usage and privacy protection.
Conclusion
RNA sequencing databases are playing an increasingly significant role in biomedical research, providing indispensable data support for gene expression studies. By leveraging these databases, researchers can gain deeper insights into the regulatory mechanisms of genes within organisms and explore molecular pathways associated with diseases. As technological advancements and data analysis tools continue to evolve, the role of RNA sequencing databases will become even more prominent. These databases will not only serve as repositories of data but also as the starting point for innovative discoveries.
Whether comprehensive databases or those focusing on specific species or biological processes, these resources are continuously evolving to offer more thorough and detailed support for scientific research. Scientists should make full use of these databases to propel new discoveries in genomics and provide novel insights for disease diagnosis and treatment.


 Sample Submission Guidelines
Sample Submission Guidelines
