High-resolution HLA Testing
Although sanger sequencing is currently the main method for HLA typing and is used in HLA tissue matching laboratories and clinical hospitals, low throughput and time consuming are the main drawbacks. Secondly, the isolation of HLA allele sequences in heterozygous specimens by first-generation sequencing also requires a more complex and expensive approach to achieve. Next-generation sequencing is rapidly becoming more widely used because of the dramatic increase in throughput and speed, and the ability to complete full-phase, high-resolution HLA typing in a single typing run.
Probe hybridization capture-based next-generation sequencing enables high-throughput and low-cost high-resolution HLA typing. The high tolerance for sequence polymorphism is an outstanding advantage of hybridization capture over PCR-based target sequence enrichment.
With the development of molecular technology and the improvement of modern medical needs, HLA typing technology has undergone the development process from serological and cytological typing with lower resolution and accuracy to genotyping with higher resolution and accuracy.
There are four common HLA genotyping:
PCR-SSP (PCR with Sequence specific primers)
PCR-SSOP(PCR with Sequence specific oligonucleotide probes)
SBT (Sequencing based typing)
NGS (Next-Generation Sequencing based typing)
Application of Next-generation Sequencing in HLA Typing
The application of next-generation sequencing for HLA typing is divided into 3 stages: (1) sequencing and typing of HLA polymorphic regions; (2) long-range PCR sequencing and typing of the entire HLA gene or whole exons; (3) single-molecule real-time sequencing (SMRT) with read-length advantage for direct sequencing and typing of HLA-I and (or) HLA-II. The first two stages are performed by 454 system, PGM and MiSeq/HiSeq, while the third stage is mainly achieved by Pacific Biosciences sequencer SMRT.
1. The 454 sequencing system was first used for HLA typing, and the principle is pyrophosphate sequencing. The read length has increased from 100-150 bp to 700 bp, and its main advantage is its fast sequencing speed and long read length.
The read length advantage is critical for producing high resolution, accurate results, and significantly reduces the uncertain results compared to SBT.
2. PGM sequencing is a low-cost and fast next-generation sequencing technology, but its short read length (about 400 bp) makes data analysis relatively difficult. From recent studies, although limited by the read length, PGM can still obtain high quality results in a short time, based on typing after full-length HLA sequencing or typing of conventional polymorphic exons by sequencing. The pressing issue is to improve or develop convenient data processing software.
3. The MiSeq/HiSeq sequencing system uses a reversible termination sequencing technology for sequencing by synthesis. Compared with other next-generation sequencing, HiSeq is less expensive, has higher throughput (600 G/run), and sequences rapidly. It is far superior to the SBT method in terms of throughput, cost effectiveness, resolution and typing speed. Compared with PGM, MiSeq obtains high throughput, low error rate, and shorter time.
4. SMRT has the advantages of fast speed and long output sequence, but its sequencing error rate is high.
The RSII sequencer launched in April 2013 has an average read length of 3000 bp, which can complete high-resolution typing and obtain complete HLA gene sequences. As the technology matures, SMRT may also increase the read length to 20 kb, which can properly solve the ambiguity problem in typing. In the near future, SMRT technology will revolutionize the HLA typing technology based on next-generation sequencing.
In terms of HLA typing, the 454 system was used first, Illumina's HiSeq, with its high data output and economical cost, occupies an important position in the field, MiSeq/HiSeq and PGM are currently the most widely used sequencing typing platforms, while SMRT and the 454 system have the advantage of long reads, and SMRT has the potential to take over later.
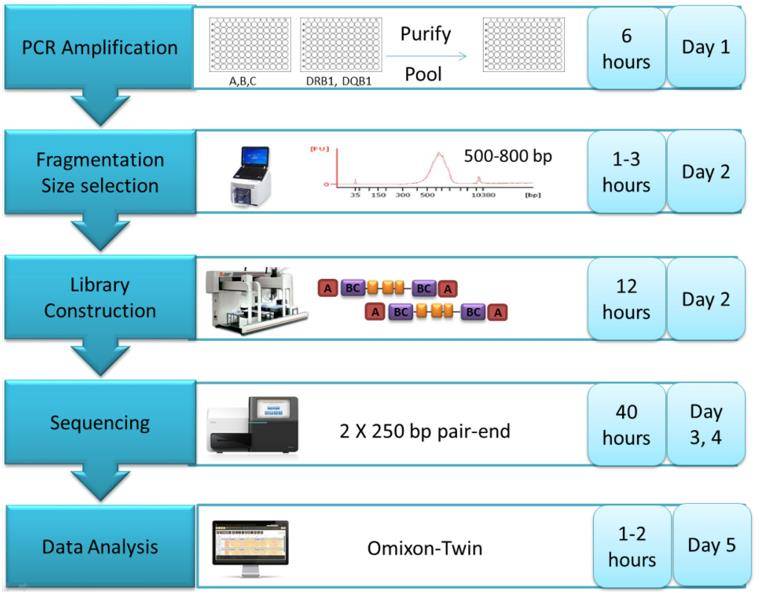 High-throughput NGS workflow (PLoS ONE 11(10) 2016).
High-throughput NGS workflow (PLoS ONE 11(10) 2016).
Characteristics of HLA typing based on next-generation sequencing technology
In addition to high throughput, short experimental cycle time and low cost, HLA typing based on next-generation sequencing technology has the following advantages:
1. High-resolution typing results
High-resolution HLA typing is essential to improve the success of transplantation and reduce the incidence of graft-versus-host disease. Before the advent of next-generation sequencing, there were two ways to obtain high-resolution results: (1) after obtaining low-resolution typing results, select appropriate probes or primers to obtain high-resolution typing; (2) after typing by SBT method, confirm the uncertain results using PCR-SSP or PCR-SSO methods to determine accurate, high-resolution typing. The next-generation sequencing method, which is similar to the SBT method, is the most accurate and direct typing method to obtain high-resolution results, and can resolve the ambiguous HLA typing results in SBT typing.
2. Solving the ambiguity problem
The percentage of HLA-A and B ambiguous results obtained based on the SBT method can reach 76.31% and 91.08%, which is an important drawback of the HLA typing gold standard-SBT method. There are two mechanisms to generate ambiguous results: (1) phase ambiguity. When the specimen HLA gene seat is heterozygous, the sequences of different allele combinations may present the results of the same typing combination. (2) The base sequences of the conventional detection region are identical, and the allelic diversity is located outside the sequencing region. The solutions include the use of SBT with PCR-SSO SSP, sequencing the region outside the conventional detection region of HLA or sequencing after separating the two alleles, but these methods are time-consuming and laborious.
HLA typing based on next-generation sequencing can effectively avoid this problem: (1) gene fragmentation amplifies and sequences individual DNA fragments, effectively obtaining the linkage phase of polymorphism; (2) massively parallel sequencing allows each round of reaction to produce a large number of exons, introns and even HLA whole gene sequencing results from different gene locations (or different specimens), which can be used to complete accurate typing.
In addition, as more and more new alleles are discovered, the variety and proportion of combinations of ambiguous results will increase, which will also drive the use of next-generation sequencing for HLA typing.
3. Whole-exome/whole-genome sequencing typing
Conventional sequencing typing focuses on HLA gene polymorphic regions, i.e., exons 2 and 3 of HLA class I genes and exon 2 of HLA class II genes. Due to cost and technical reasons, only about 10% of the named genes in the IMGT/HLA database have HLA full gene sequences.
With the maturation of HLA typing methods by next-generation sequencing and the reduction of sequencing costs, HLA whole-exome or whole-genome sequencing typing has been increasingly reported. This method is beneficial for discovering new HLA alleles, resolving ambiguities, and obtaining ultra-high resolution, which can enrich the data of IMGT/HLA database and marrow transplantation, and facilitate the study of HLA polymorphism and molecular evolution in the population, and provide great convenience for future scientific research and clinical work.
The shortcomings of HLA typing by next-generation sequencing
However, before this application is popularized, the following obstacles remain: (1) although the performance of the next-generation sequencing method has improved significantly compared with other methods, it is undoubtedly a labor-intensive, material and financial project to obtain a large-scale population HLA whole-genome sequence; (2) the operational specifications of HLA whole-genome or whole-exome sequencing typing by next-generation sequencing need to be gradually improved; (3) whole-genome sequencing typing is bound to discover a large number of new HLA alleles, and issues such as their naming and confirmation as well as the integration and sharing of massive information need to be considered.
The emergence of new technologies, along with obvious advantages, also has certain limitations. The shortcomings of HLA typing by next-generation sequencing mainly lie in: (1) short read length, which is the main bottleneck of next-generation sequencing technology and brings certain difficulties to the later genome assembly, it is gratifying that SMRT read length can still be completed efficiently for HLA typing; (2) high data analysis and software requirements, a series of excellent data processing software has emerged accordingly, such as Omixon Target HLAE201, NGSengine, HLAreporter, Op-tiType, etc.
Conclusion
As a classical HLA genotyping technology, PCR-SBT/SSP/SSO is still the mainstream method internationally, but HLA typing by next-generation sequencing has obvious advantages in terms of high throughput, high resolution, resolution of ambiguous typing results and discovery of new alleles. With the development of technology and popularization of application, large sample size control experiments under different next-generation sequencing platforms; naming system improvement and information sharing platform establishment; data processing software development and evaluation as well as standardized operation protocol development deserve further attention in future research. Due to the complexity of HLA molecules, the description of HLA diversity in the population and the exploration of evolutionary mechanisms appear to be relatively difficult. HLA typing studies based on next-generation sequencing provide convenience for the establishment of large-scale bone marrow libraries, high-efficiency matching work, and the study of HLA gene structure and molecular function, which will lead to the further development of HLA research and applications.
References:
- Trowsdale J,Knight JC.Major histocompatibility complex genomics and human disease. Annu Rev Genomics, Hum Genet,2013,14:301-323.
- Mosaad YM.Clinical role of human leukocyte antigen in health and disease.Scand J Immunol,2015,82(4):283-306.
- Yamamoto F,Hoglund B,Fernandez-Vina M,et al.Very high resolution single pass HLA genotyping using ampli-con sequencing on the 454 next Generation DNA sequenc-ers:Comparison with Sanger sequencing.Hum Immu-nol,2015,76(12).910-916.
- Barone JC,Saito K,Beutner K,et al.HLA-genotyping of clinical specimens using lon Torrent-based NGS.Hum Immunol,2015,76(12):903-909.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 High-throughput NGS workflow (PLoS ONE 11(10) 2016).
High-throughput NGS workflow (PLoS ONE 11(10) 2016).