CD Genomics provides the exosome researchers with a comprehensive service to investigate exosome-associated RNA information.
The Introduction of Exosomal RNA Sequencing
Exosomes are cell-derived vesicles that are present in many and perhaps all eukaryotic fluids, including blood, urine, and cultured medium of cell cultures. Once released, exosomes travel throughout the body, fuse with other cells and transfer their cargo to the acceptor cell. Recent research has uncovered evidence that exosomes play an important role in intercellular communication and disease transmission.
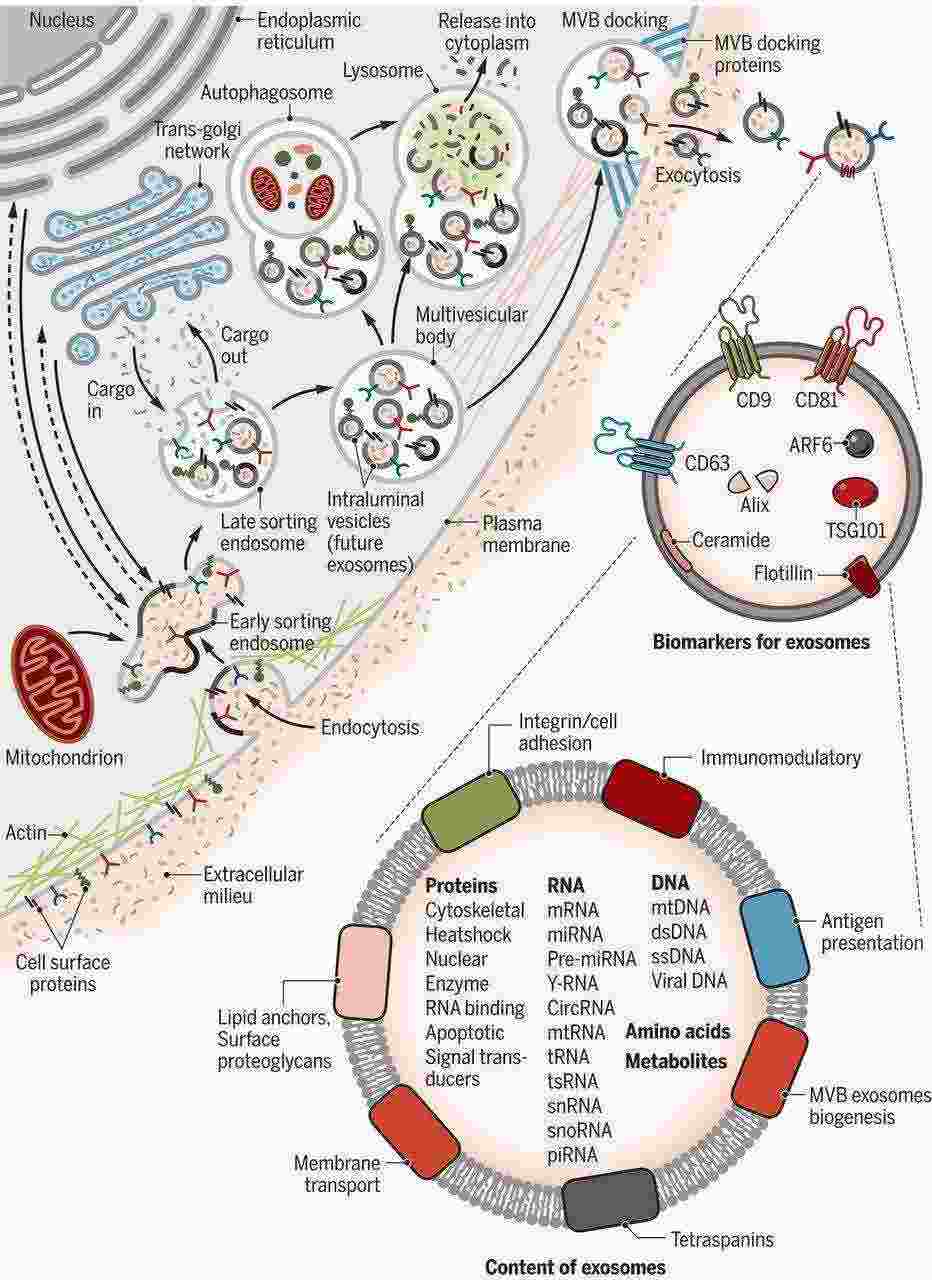 Figure 1. Biogenesis and identification of exosomes. (Kalluri et al., 2020)
Figure 1. Biogenesis and identification of exosomes. (Kalluri et al., 2020)
CD Genomics provides customers with the comprehensive exosomal RNA sequencing service. Exosomes contain distinct subsets of microRNAs, mRNAs, lncRNAs and other ncRNAs. Evidence is accumulating that the delivery of miRNA via exosomal transmission can result in fully functional or translatable long RNA within a recipient cell. Exosomal miRNAs may have important functions in cell-cell communication and have potential as biomarkers to detect and monitor disease. Exosomal RNA profiling provides insights into the exosome-associated RNA function and enables the applications as shown below.
- Identifying changes in exosomal miRNA under disease states is considered to be a reliable method to discover sequence signatures for the diagnosis and prognosis of disease
- The identity and expression level of exosomal mRNA have been considered a valuable approach to discover biomarkers from blood-based, saliva, and urine biofluid samples
- Long noncoding RNAs (lncRNA) are also present in exosomes and can show expression patterns that are distinct from the cellular level
Advantages of Exosomal RNA Sequencing
- Non-invasive Sampling: RNA samples are obtained from extracellular vesicles in bodily fluids such as blood, urine, and saliva, eliminating the need for invasive procedures. This approach reduces patient discomfort, minimizes risks, and enhances patient compliance.
- Robust Stability: RNA enclosed within extracellular vesicles is shielded by vesicle membranes, rendering it more stable compared to free-floating RNA, thus less susceptible to degradation by RNA enzymes. This heightened stability streamlines sample handling and storage, ultimately bolstering data reliability.
- Diversity and Richness: Exosomal RNA encompasses various RNA molecule types including mRNA, miRNA, and lncRNA, furnishing a wealth of genetic expression data that mirrors authentic cellular physiological states and pathological changes.
- Real-time Cellular Reflection: Originating from living cells, extracellular vesicles offer real-time insights into cellular physiological and pathological conditions. Consequently, exosomal RNA sequencing can be harnessed for monitoring disease onset, progression, and treatment outcomes.
- High Sensitivity and Specificity: Exosomal RNA sequencing techniques boast remarkable sensitivity, capable of detecting low-abundance RNA molecules. Additionally, the vesicles' specificity stemming from diverse cell origins enables early disease diagnosis and precise subtyping through RNA sequencing.
- Wide-ranging Applications: Exosomal RNA Sequencing holds vast potential across diverse disease studies encompassing cancer, neurodegenerative disorders, cardiovascular diseases, and beyond. Its utility spans early disease diagnosis, prognosis assessment, treatment monitoring, and the discovery of novel biomarkers.
- Facilitating Personalized Healthcare: Through exosomal RNA sequencing, personalized gene expression profiles can be garnered. When integrated with other molecular information, this can pave the way for tailoring individualized treatment strategies, enhancing therapeutic efficacy, and minimizing adverse reactions.
Exosomal RNA Sequencing Workflow
CD Genomics provides the end-to-end exosomal RNA NGS service including isolation of exosomes from serum, plasma, urine, tissue culture media, or other samples and extraction of their associated RNA using commercial kits. CD Genomics' exosomal NGS service combines Illumina NGS barcoded libraries and next-generation sequencing platforms, offering deep sequencing data of the highest quality to accelerate your exosomal RNA research.
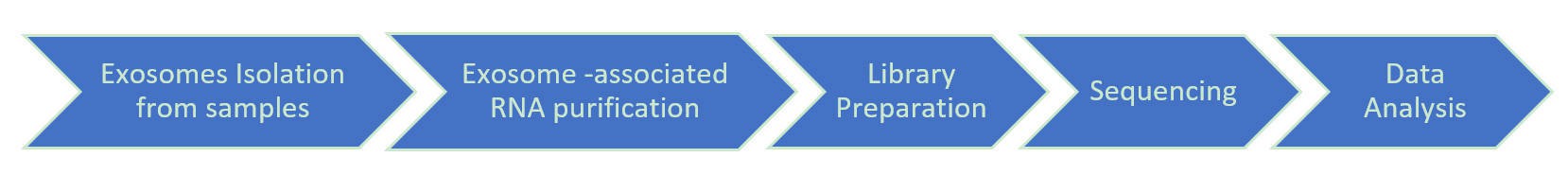
Service Specification
Sample Requirements
|
|
Click |
Sequencing Strategies
|
| Data Analysis We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
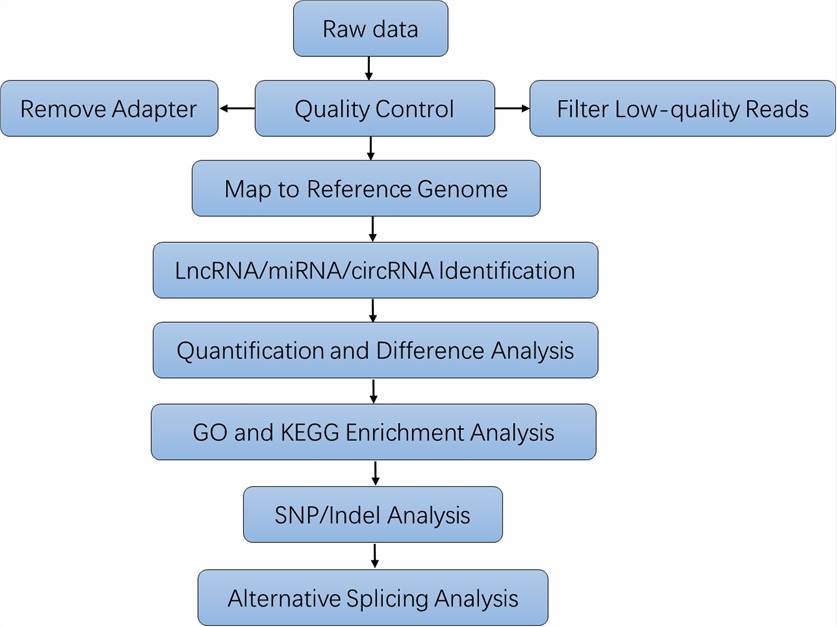
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in exosomal RNA sequencing for your writing (customization)
CD Genomics' Exosomal RNA sequencing conference focuses on the links between cell variation in tissues and organ function and further elucidates the origins of diseases. If you have additional requirements or questions, please feel free to contact us.
Reference:
- Kalluri R, LeBleu V S. The biology, function, and biomedical applications of exosomes. Science, 2020, 367(6478): eaau6977.
Partial results are shown below:

Sequencing quality distribution

A/T/G/C Distribution

IGV Browser Interface

Correlation Analysis Between Samples

PCA Score Plot

Venn Diagram

Volcano Plot

Statistics Results of GO Annotation

KEGG Classification
1. What is the function of the exosome in RNA?
The exosome functions as a pivotal player in RNA metabolism, intricately involved in RNA degradation and processing mechanisms. Operating as an RNA exoribonuclease complex within eukaryotic cells, the exosome is dedicated to the surveillance and breakdown of a spectrum of RNA species. This pivotal role is paramount for sustaining the equilibrium of cellular RNA, orchestrating gene expression patterns, and orchestrating responses to cellular stressors. Additionally, exosomes partake in RNA surveillance pathways, meticulously overseeing the quality assurance of RNA transcripts pre-translation.
2. What RNA species are in exosomes?
As for the RNA species encapsulated within exosomes, they encompass a rich assortment, encompassing messenger RNA (mRNA), microRNA (miRNA), long non-coding RNA (lncRNA), transfer RNA (tRNA), and various minor non-coding RNAs. Safeguarded by the lipid bilayer membrane of the vesicles, these RNA molecules reside within exosomes shielded from rapid degradation. The presence of particular RNA species within exosomes is contingent upon variables such as cell type, physiological milieu, and pathological states. Significantly, these exosomal RNAs have the capacity to be transferred to recipient cells, where they wield influence over gene expression patterns and cellular functions within both normal and pathological contexts.
3. Which library preparation method should we choose: low-input or standard?
When the extracted RNA quantity exceeds 100ng, it is advisable to employ the standard rRNA-depletion method, utilizing 100ng of RNA for total transcriptome library preparation. Generally, a larger input of template during library preparation results in higher coverage of genetic information. The low-input library preparation approach is utilized when the sample quantity is limited; however, whenever feasible, conventional library preparation methods are preferred for experimental procedures.
4. Regarding exosome studies, should we use plasma or serum?
The recommendation leans towards using plasma due to the propensity of platelet activation during clotting in serum collection, leading to the generation of a significant number of exosomes and other vesicles. Consequently, the vesicles obtained from serum are consistently greater in number compared to those in plasma, with over 50% of vesicles originating from platelets. Thus, plasma serves as a superior medium for investigating exosomes in pathological physiological conditions. The majority of circulating exosome research samples make use of plasma.
5. What are the key considerations for exosome RNA sequencing?
In the context of exosome RNA sequencing, several critical considerations merit attention:
Sample Quality: Ensuring the quality of the sample source and adherence to standardized collection procedures.
Exosome Purity: Implementing efficient extraction methods to ensure the purity and integrity of exosomes.
RNA Quality: Extracted RNA should exhibit excellent integrity and purity to prevent degradation.
Technological Platform: Selection of an appropriate sequencing platform to guarantee data accuracy and reliability.
Data Analysis: Engaging a proficient bioinformatics analysis team to ensure the scientific rigor and robustness of data analysis.
6. What is the method of library construction?
CD Genomics typically prepares stranded, rRNA-depleted cDNA libraries from the DNA-free total RNA.
Cooked pork-derived exosome nanovesicles mediate metabolic disorder—microRNA could be the culprit
Journal: Journal of Nanobiotechnology
Impact factor: 11.509
Published: 09 March 2023
Abstract
Extracellular vesicles (EVs), including exosomes and ectosomes, represent lipid-enclosed structures released by all cell types. Exosomes, typically ranging from 40 to 160 nm in diameter, act as crucial mediators of intercellular communication and hold promise as diagnostic markers for diseases like cancer, Parkinson's, and Alzheimer's. Encompassing lipids, proteins, and microRNAs (miRNAs), exosomal miRNAs derived from dietary sources can be absorbed and exert pivotal biological functions within organisms. This investigation endeavors to characterize exosomes and miRNA profiles extracted from cooked pork utilizing high-throughput sequencing, elucidating their impact on murine physiology. It highlights the resilience of exosomes and miRNAs under elevated thermal conditions and underscores the dietary significance of pork as a primary nutritional component.
Materials & Methods
- Cooked porcine muscle
- Fat and liver samples
- Exosomes isolation
- RNA extraction
- Library construction
- RNA-Seq
- RT-qPCR
- DEGs analysis
- miRNA target gene prediction
- Gene Ontology (GO) analysis
- KEGG enrichment analysis
- Protein–protein interaction (PPI) networks
Results
The research team initially subjected various pork tissues (adipose tissue, muscle, liver) to thorough cooking in water, followed by the extraction of their exosomes using differential ultracentrifugation. Examination of the extract under transmission electron microscopy (TEM) and atomic force microscopy (AFM) revealed exosome-like nanovesicles. Further analysis using dynamic light scattering (DLS) indicated that 80% of the particles ranged from 20-200nm, with an average size of 70.29nm. Subsequent flow cytometry revealed positive rates of 84.5% for the exosome-specific markers CD63 and 95.9% for CD81. These characterization findings confirm the successful extraction of exosomes from cooked pork tissues.
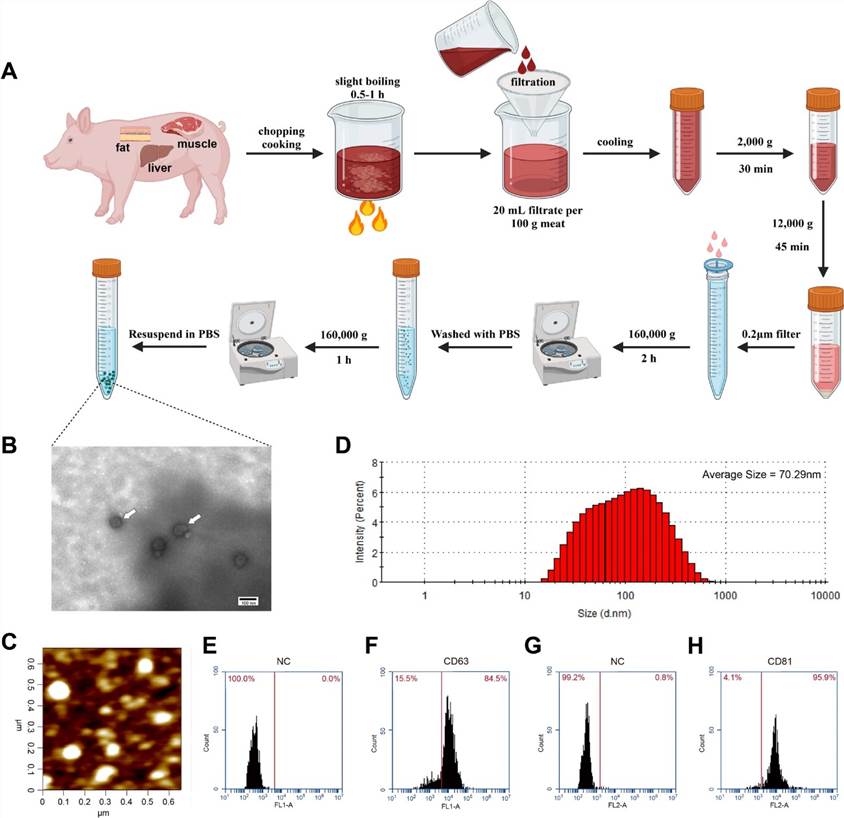 Fig 1. Identification of exosomes isolated from cooked meat.
Fig 1. Identification of exosomes isolated from cooked meat.
Upon high-throughput sequencing analysis, it was observed that exosomes derived from different tissues exhibit variations in the types and quantities of miRNAs they contain. In the exosomal content from cooked pork muscle (PMEx), miR-1, miR-133a-3p, and miR-206 collectively constitute 80% of the total miRNAs. Interestingly, in exosomes from cooked pork liver (PLEx), miR-122 predominates, representing 37.8% of the total miRNAs, while in exosomes from cooked pork fat tissue (PFEx), miR-125b is notably abundant.
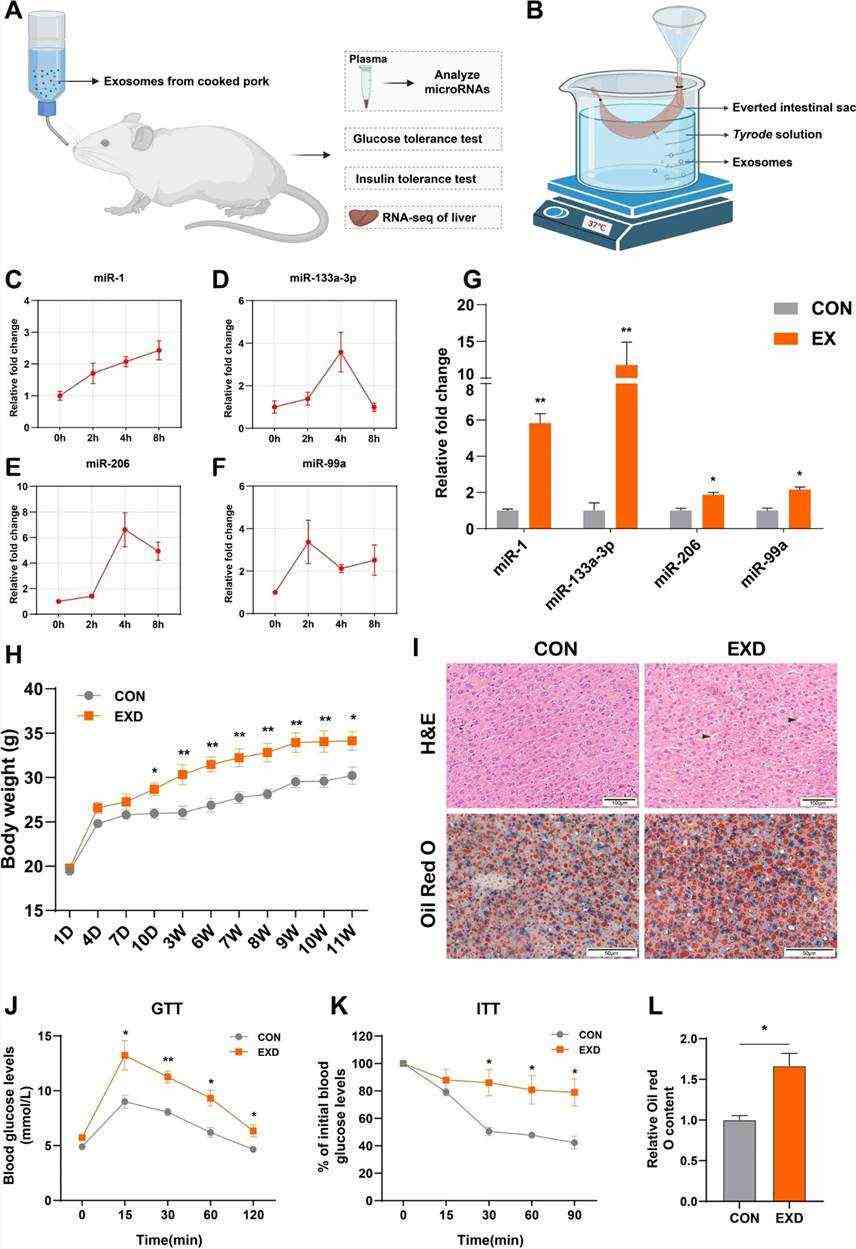 Fig 2. Mice were given drinking water supplemented with PME.
Fig 2. Mice were given drinking water supplemented with PME.
After feeding the extracted exosomes to mice for a period, several notable observations were made: (1) a significant increase in body weight was observed; (2) levels of miR-1, miR-133a-3p, miR-206, and miR-99a in the blood markedly increased, with sustained elevation observed during the intestinal villus inversion experiment; (3) the presence of vacuoles and lipid droplets in liver tissues; (4) fasting glucose levels significantly rose and the consumption rate slowed after glucose injection on an empty stomach. The experimental outcomes confirm that CM-Exo can be absorbed by the intestines into the bodies of mice, influencing body weight, blood miRNA levels, and participating in biological regulatory processes such as glucose tolerance, insulin resistance, and liver lipid metabolism.
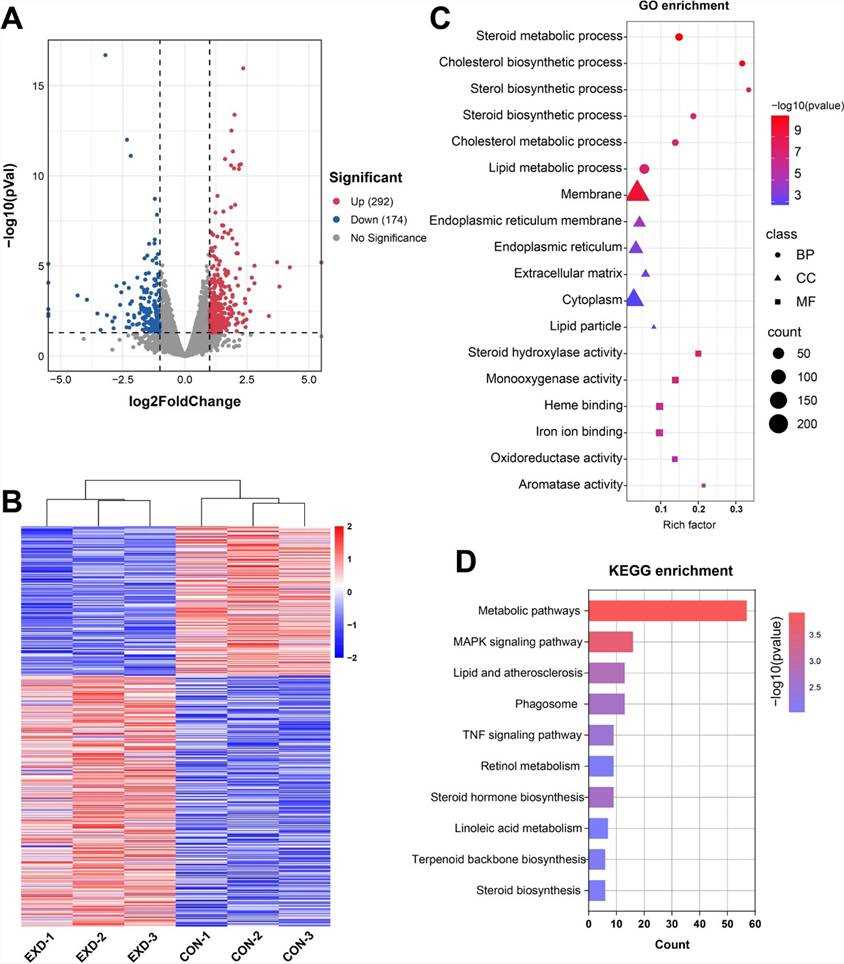 Fig 3. The overview of the transcriptomic changes of mouse liver.
Fig 3. The overview of the transcriptomic changes of mouse liver.
Transcriptome analysis of mouse liver samples identified 446 differentially expressed genes (DEGs), with 292 upregulated and 174 downregulated genes. GO and KEGG enrichment analyses indicated that these DEGs are predominantly involved in steroid metabolism. Subsequently, a putative miRNA-mRNA-lipid metabolism regulatory network was constructed, revealing a significant interconnection between exosomal miRNA and hepatic lipid metabolic pathways.
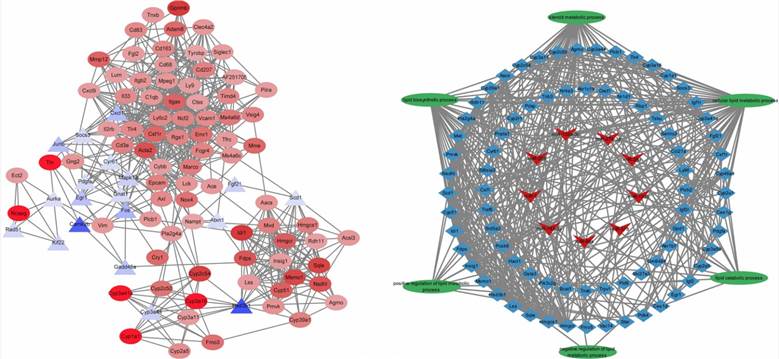 Fig 4. Top 100 hub genes identified from the PPI network; miRNA–mRNA–lipid metabolism pathways regulatroy network.
Fig 4. Top 100 hub genes identified from the PPI network; miRNA–mRNA–lipid metabolism pathways regulatroy network.
Conclusion
Extracellular vesicles, or exosomes, have garnered significant attention in cellular biology due to their potential therapeutic and diagnostic applications. Initially viewed as mere cellular waste, we now understand that their functions extend far beyond waste disposal. Exosomes represent a novel mode of cellular communication, contributing to a range of biological processes relevant to health and disease.
Using differential ultracentrifugation, the research team successfully isolated exosomes from cooked pork (adipose tissue, muscle, liver). Transcriptomic analysis revealed distinct differences in the miRNA content encapsulated within exosomes sourced from different tissues. Feeding exosomes to mice and performing intestinal loop experiments confirmed that CM-Exo can be absorbed by the intestinal tract, impacting body weight and blood miRNA levels, leading to insulin resistance and disturbances in hepatic lipid metabolism. Overall, microRNAs within CM-Exo likely serve as key regulatory factors in inducing metabolic disruptions in mice.
Reference:
- Shen L, Ma J, Yang Y, et al. Cooked pork-derived exosome nanovesicles mediate metabolic disorder—microRNA could be the culprit. Journal of Nanobiotechnology, 2023, 21(1): 83.
Here are some publications that have been successfully published using our services or other related services:
Chaperone-Mediated Autophagy Controls Proteomic and Transcriptomic Pathways to Maintain Glioma Stem Cell Activity
Journal: Cancer research
Year: 2022
Circular DNA tumor viruses make circular RNAs
Journal: Proceedings of the National Academy of Sciences
Year: 2018
Repeated immunization with ATRA-containing liposomal adjuvant transdifferentiates Th17 cells to a Tr1-like phenotype
Journal: Journal of Autoimmunity
Year: 2024
Role of the histone variant H2A.Z.1 in memory, transcription, and alternative splicing is mediated by lysine modification
Journal: Neuropsychopharmacology
Year: 2024
FAK loss reduces BRAFV600E-induced ERK phosphorylation to promote intestinal stemness and cecal tumor formation
Journal: Elife
Year: 2023
Identification of circular RNAs regulating cardiomyocyte proliferation in neonatal pig hearts
Journal: JCI insight
Year: 2024
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
