Symbiosis encompasses the intricate, interdependent relationships between species in the natural world. Harmonious coexistence represents a sustainable and mutually beneficial bond among species, exemplified by the symbiotic partnership between rhizobia and specific crops. As human beings, our quest for harmony extends beyond our external environment; it also entails fostering a balanced relationship with the microorganisms residing within our bodies, known as the intestinal flora. This microbial community plays a pivotal role in the daily processing of our dietary intake, transforming carbohydrates and other energy-rich macromolecules into essential molecular components for our use. Consequently, the metabolic activities of the intestinal flora, particularly concerning various energy substrates, hold direct implications for human health.
But what influence does the gut flora exert on insulin resistance, and through what mechanisms?
A recent publication by Hiroshi Ohno's research team at the Laboratory of Intestinal Ecosystems, RIKEN Center for Integrative Medicine in Yokohama, Japan, sheds light on this inquiry. In their article, titled "Gut Microbial Carbohydrate Metabolism's Impact on Insulin Resistance," published in Nature, the team delves into their exploration of the effects of intestinal flora on insulin resistance. Their study employed metagenomic, lipidomic, and transcriptomic data analyses to unveil a remarkable phenomenon: the intestinal flora, specifically Mycobacterium spp., engages in the metabolism of carbohydrates, with a focus on monosaccharides like glucose, fructose, galactose, mannose, xylose, and arabinose. Astonishingly, these metabolic activities of the intestinal flora contribute to the amelioration of insulin resistance in human beings.
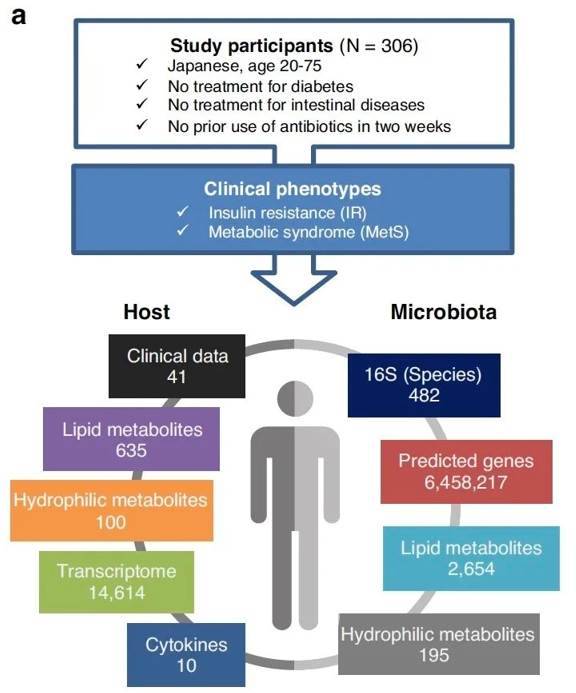 Overview of multi-omics analysis and data. (Takeuchi et al., 2023)
Overview of multi-omics analysis and data. (Takeuchi et al., 2023)
A Comprehensive Analysis of Fecal Metabolites in Understanding Insulin Resistance Pathways
The researchers meticulously gathered fecal and blood samples from willing volunteers, subjecting them to a comprehensive metabolomic analysis utilizing two advanced mass spectrometry platforms. This exhaustive process revealed a remarkable array of findings, with 195 hydrophilic metabolites and 2,654 lipid metabolites detected within the fecal samples, and 100 hydrophilic metabolites and 653 lipid metabolites within the blood samples.
In order to comprehensively evaluate the variations in gut flora function, the researchers categorized fecal metabolites and predicted genes into coherent groups known as co-abundance groups (CAGs). Furthermore, they conducted KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analyses to deepen their insights.
To assess the capacity of fecal sample histology data to accurately predict insulin resistance, the researchers embarked on a robust investigation. Based on the Random Forest algorithm, they initiated by comparing the area under the curve (AUC) of the receiver operating characteristic (ROC) curve, a crucial tool in diagnostic accuracy assessment.
For the model's predictor variables, a meticulous selection process was executed. The minimum redundancy maximum correlation algorithm was employed to curate PCR data from the 16S rRNA V1-V2 hypervariable region of fecal flora, the metabolome, the metagenome set, and their combined datasets. Notably, the study results established that fecal metabolite data exhibited strong potential for illuminating the intricacies of insulin resistance pathology.
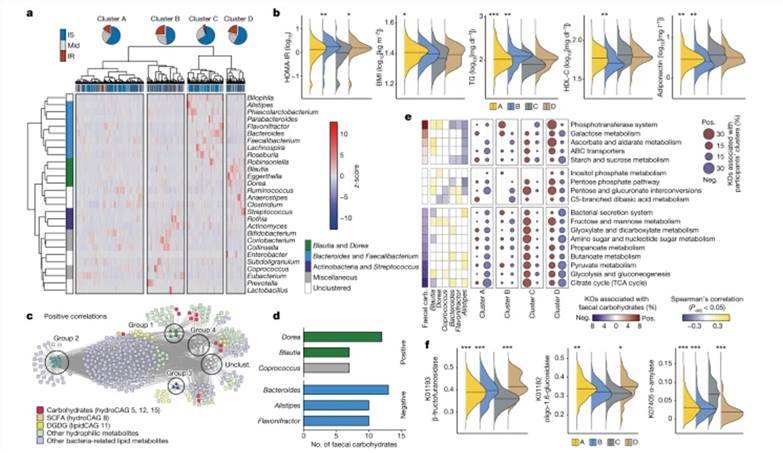 IR-associated fecal metabolites are associated with altered gut microbiota and microbial genetic functions. (Takeuchi et al., 2023)
IR-associated fecal metabolites are associated with altered gut microbiota and microbial genetic functions. (Takeuchi et al., 2023)
Following these initial steps, the research team promptly conducted an extensive data analysis. Their investigation revealed a notable surge in fecal carbohydrate metabolites among insulin-resistant individuals. This surge was particularly pronounced in the case of monosaccharides like glucose, fructose, galactose, mannose, xylose, and arabinose, which exhibited marked elevation in the fecal samples of individuals with insulin resistance.
Moreover, the researchers also uncovered the presence of ester metabolites associated with insulin resistance within co-abundance group 11 (CAG11). This group included lysophospholipids, bile acids, esteroylcarnitine, and digalactosyl/glucosyldiacylglycerol (DGDG), compounds that featured glucose and/or galactose components within their structural makeup. Interestingly, within co-abundance group 41 (CAG41), the scientists noted a lack of association between insulin resistance and DGDGs with varying acyl chains. This observation hinted at the possibility that esters with diverse acyl chains may fulfill distinct physiological roles.
Subsequently, the researchers delved into an examination of the interplay between bacterial flora and metabolites within insulin-resistant individuals. Utilizing 16S rRNA sequencing data, they identified four major groups in the volunteers' fecal samples: Lachnospiraceae, Bacteroides, Actinobacteria, and unclassifiable bacteria. A deeper analysis illuminated a positive correlation between Lachnospiraceae and unclassifiable bacteria and insulin resistance, while Bacteroides exhibited differential behavior, particularly in their capacity to metabolize carbohydrates within a strain of the genus Alistipes indistinctus (A. indistinctus).
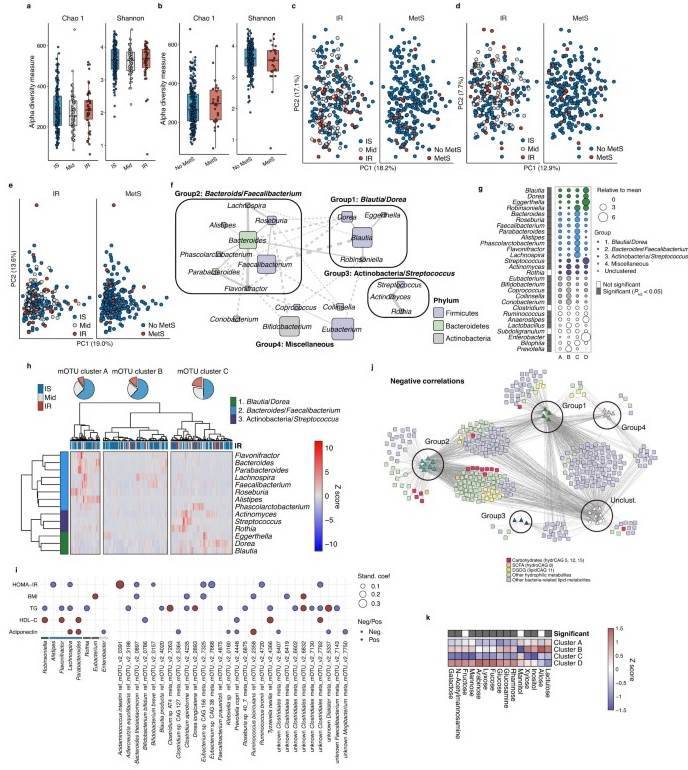 Fecal microbiota in IR. (Takeuchi et al., 2023)
Fecal microbiota in IR. (Takeuchi et al., 2023)
The researchers also scrutinized inflammation data, uncovering a pivotal role played by cytokines such as IL-10, serpin E1, adiponectin, and leptin in mediating the connection between fecal carbohydrates and host insulin resistance markers, including the homeostatic model assessment of insulin resistance (HOMA-IR). It is worth noting that most of these causal relationships were established through sophisticated in silico models.
In conclusion, the researchers conducted comprehensive investigations to establish a definitive causal link between gut flora, fecal carbohydrates, and metabolic diseases in a mouse model. Initially, they scrutinized metabolites within cultures comprising 22 different strains, each associated with either insulin sensitivity (IS) or resistance. The data revealed that anabolic species, such as A. indistinctus, Alistipes finegoldii, and Alistipes putredinis, exhibited a pronounced preference for monosaccharide carbohydrates like glucose, mannose, and glucosamine. Notably, the strain A. indistinctus outperformed the rest, displaying remarkable versatility in carbohydrate consumption.
Subsequently, seven strains associated with insulin sensitivity were chosen for implantation into mice to assess their therapeutic potential against metabolic diseases. The outcomes demonstrated substantial improvements in the metabolic status of mice following the introduction of the A. indistinctus strain. These results consistently mirrored the earlier findings, confirming that mice implanted with A. indistinctus exhibited the most favorable metabolic outcomes.
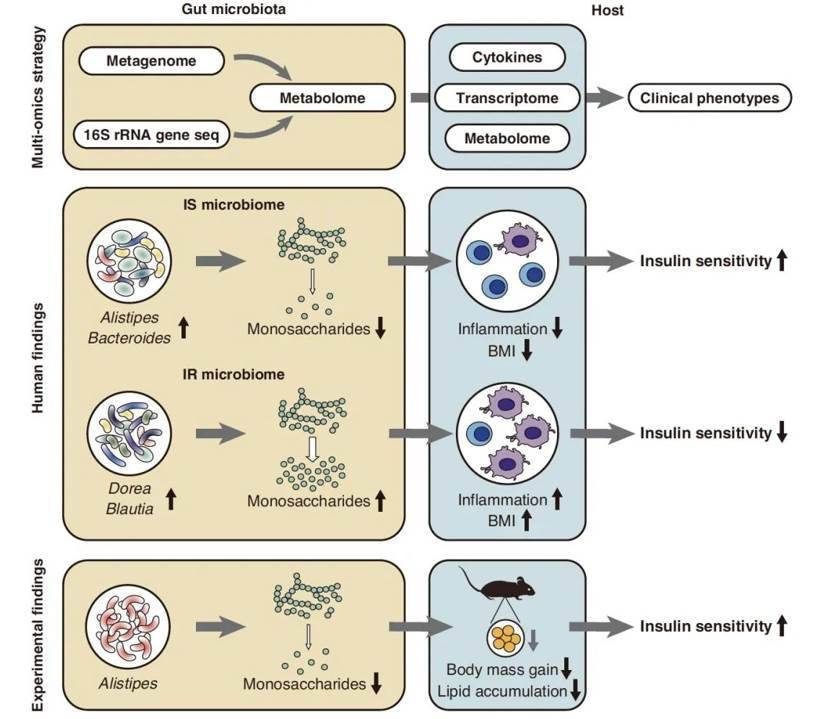 10 Alistipes indistinctus reduces intestinal carbohydrates. (Takeuchi et al., 2023)
10 Alistipes indistinctus reduces intestinal carbohydrates. (Takeuchi et al., 2023)
Reference:
-
Takeuchi, Tadashi, et al. "Gut microbial carbohydrate metabolism contributes to insulin resistance." Nature (2023): 1-7.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Overview of multi-omics analysis and data. (Takeuchi et al., 2023)
Overview of multi-omics analysis and data. (Takeuchi et al., 2023) IR-associated fecal metabolites are associated with altered gut microbiota and microbial genetic functions. (Takeuchi et al., 2023)
IR-associated fecal metabolites are associated with altered gut microbiota and microbial genetic functions. (Takeuchi et al., 2023) Fecal microbiota in IR. (Takeuchi et al., 2023)
Fecal microbiota in IR. (Takeuchi et al., 2023) 10 Alistipes indistinctus reduces intestinal carbohydrates. (Takeuchi et al., 2023)
10 Alistipes indistinctus reduces intestinal carbohydrates. (Takeuchi et al., 2023)