To synchronize with the rhythmic alterations in the environment caused by the Earth's rotation, higher plants have developed an internal timekeeping mechanism known as the biological clock. This biological clock accurately regulates various biological processes within organisms, including gene expression, cellular metabolism, and growth and development. This orchestration ensures optimal energy acquisition and utilization. In the context of rice, approximately one-third of the genes that are actively transcribed exhibit cyclic patterns. However, the intricate mechanisms through which the biological clock governs the rhythmic expression of genes remain insufficiently examined. Furthermore, the epigenomic and three-dimensional genomic foundations of rhythmic gene expression remain unclear.
Researchers have assembled extensive genome-wide datasets encompassing RNA-seq, H3K9ac ChIP-seq, ATAC-seq, and BL-Hi-C data across three distinct light conditions. Through their efforts, they have unveiled the circadian oscillation motif associated with the histone H3K9ac modification and the dynamic rhythmic alterations in the 3D structure of the genome. Additionally, they have delved into the variations in expression of rhythmic rice genes in response to diverse light conditions.
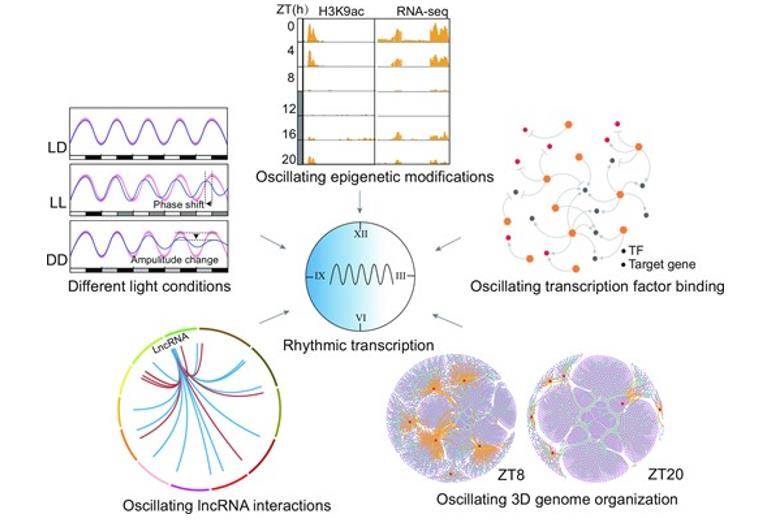 Integrated 3D genome, epigenome and transcriptome analyses reveal transcriptional coordination of circadian rhythm in rice. (Zhang et al., 2023)
Integrated 3D genome, epigenome and transcriptome analyses reveal transcriptional coordination of circadian rhythm in rice. (Zhang et al., 2023)
Rhythmic Changes in H3K9ac Modification Sites
The modification sites of H3K9ac exhibited noticeable rhythmic alterations under varying conditions. It was determined that the extent of H3K9ac modification signals displayed a notably positive correlation with the degree of rhythmic changes in expression at the corresponding sites. The summit of histone acetylation modification in terms of expression took precedence over the peak of rhythmic gene expression for a span of 1-2 hours. This suggests that the rhythmic alterations in H3 acetylation play a crucial role in propelling the transcriptional expression of genes exhibiting rhythmic patterns. Furthermore, the study revealed significant variations in the oscillatory characteristics of rhythmic gene expression in rice under distinct light conditions. Within the scope of genome-wide rhythmic gene expression, the expression levels were lower under low light (LL) conditions compared to long-day (LD) conditions, but higher than those under continuous darkness (DD) conditions. The amplitude of expression under LL conditions was lower than that in LD conditions, yet higher than that in DD conditions. Additionally, the phase was advanced under LL conditions and delayed under DD conditions. The examination of well-known core biological clock genes' expression patterns under various conditions confirmed their alignment with the expression patterns of rhythmic genes across the entire genome. Continuous darkness induced a phase delay in rhythmic gene expression, whereas continuous light resulted in reduced amplitude of rhythmic gene expression.
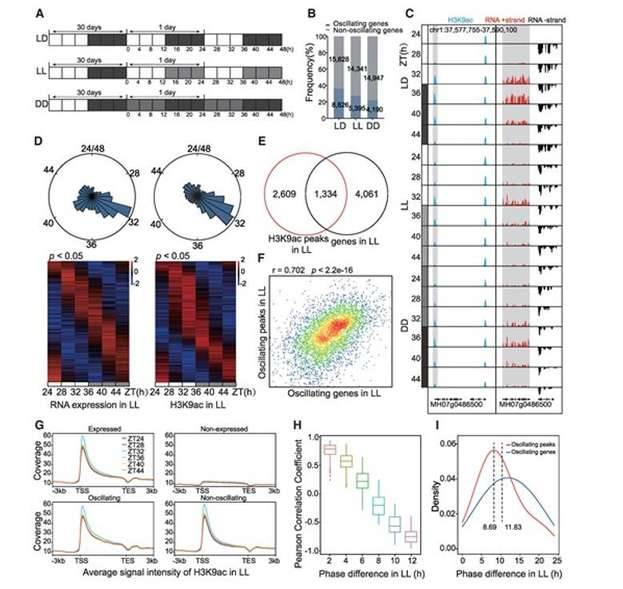 Circadian changes in H3 acetylation regulates oscillating gene expression. (Zhang et al., 2023)
Circadian changes in H3 acetylation regulates oscillating gene expression. (Zhang et al., 2023)
Dynamics of Transcription Factor Binding and Regulatory Networks in Circadian Gene Expression
Examination of the ATAC-seq data exposed that accessible chromatin areas remained relatively stable throughout the 24-hour day cycle. However, a substantial quantity of transcription factors exhibited time-dependent binding to these chromatin regions, orchestrating the rhythmic expression of genes. Subsequent to this, scientists charted the regulatory network formed by these oscillatory binding transcription factors, connecting them to genes that displayed rhythmic expression patterns. This connection was established by incorporating the rhythmic footprints found in open chromatin regions of fundamental biological clock genes and employing predictive modeling for transcription factors. Notably, the investigation highlighted the significant roles played by transcription factors related to light and temperature signaling in driving circadian rhythm fluctuations and coordinating the expression of crucial biological clock genes.
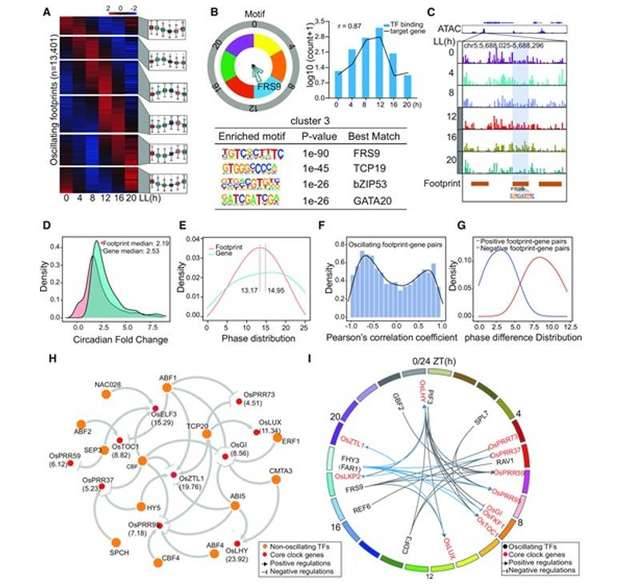 Mapping the regulatory network of oscillating transcription factors. (Zhang et al., 2023)
Mapping the regulatory network of oscillating transcription factors. (Zhang et al., 2023)
Summary
Using in situ Hi-C technology, researchers conducted a comparative analysis of chromatin structure at two time points: ZT8 and ZT20. The genes exhibiting time-specific rhythmic expression at ZT8 were observed to be densely packed, while at ZT20, they appeared to be more loosely arranged. This observation suggests that the rhythmic expression of these genes is synchronized with the changing 3D structure of chromatin throughout the day. Furthermore, certain rhythmic long non-coding RNAs (lncRNAs) displayed synchronized oscillatory H3K9ac modifications, and these lncRNAs were co-expressed with neighboring rhythmic genes. This co-expression is believed to stem from interactions within the chromatin space. These findings shed light on a potential mechanism involving the alignment of oscillating H3K9ac modifications, dynamically bound transcription factors, and co-expressed rhythmic lncRNAs and genes. Ultimately, these insights offer a fresh perspective on the dynamic alterations in spatial chromatin conformation that are linked to the rhythmic expression of genes.
Reference:
-
Zhang, Ying, et al. "Integrated 3D genome, epigenome and transcriptome analyses reveal transcriptional coordination of circadian rhythm in rice." Nucleic Acids Research (2023): gkad658.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Integrated 3D genome, epigenome and transcriptome analyses reveal transcriptional coordination of circadian rhythm in rice. (Zhang et al., 2023)
Integrated 3D genome, epigenome and transcriptome analyses reveal transcriptional coordination of circadian rhythm in rice. (Zhang et al., 2023) Circadian changes in H3 acetylation regulates oscillating gene expression. (Zhang et al., 2023)
Circadian changes in H3 acetylation regulates oscillating gene expression. (Zhang et al., 2023) Mapping the regulatory network of oscillating transcription factors. (Zhang et al., 2023)
Mapping the regulatory network of oscillating transcription factors. (Zhang et al., 2023)